Direct readout of heterochromatic H3K9me3 regulates DNMT1-mediated maintenance DNA methylation.
Ren, W., Fan, H., Grimm, S.A., Guo, Y., Kim, J.J., Yin, J., Li, L., Petell, C.J., Tan, X.F., Zhang, Z.M., Coan, J.P., Gao, L., Cai, L., Detrick, B., Cetin, B., Cui, Q., Strahl, B.D., Gozani, O., Wang, Y., Miller, K.M., O'Leary, S.E., Wade, P.A., Patel, D.J., Wang, G.G., Song, J.(2020) Proc Natl Acad Sci U S A 117: 18439-18447
- PubMed: 32675241
- DOI: https://doi.org/10.1073/pnas.2009316117
- Primary Citation of Related Structures:
6PZV - PubMed Abstract:
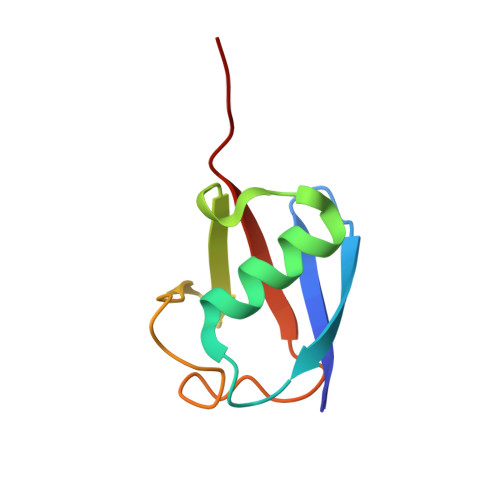
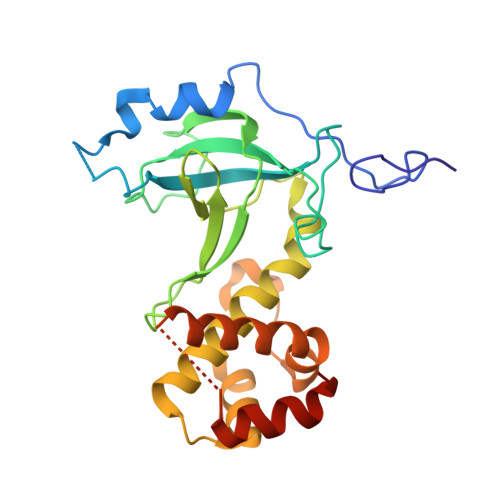
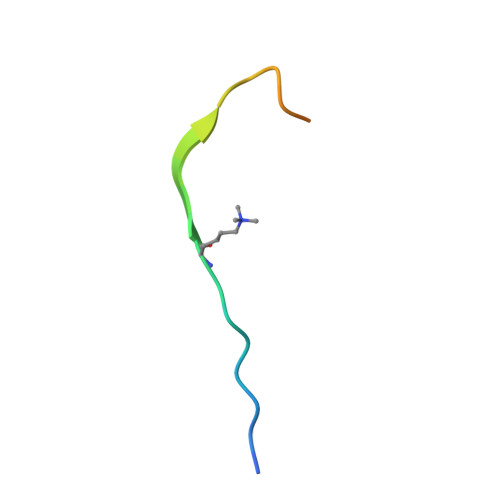
In mammals, repressive histone modifications such as trimethylation of histone H3 Lys9 (H3K9me3), frequently coexist with DNA methylation, producing a more stable and silenced chromatin state. However, it remains elusive how these epigenetic modifications crosstalk. Here, through structural and biochemical characterizations, we identified the replication foci targeting sequence (RFTS) domain of maintenance DNA methyltransferase DNMT1, a module known to bind the ubiquitylated H3 (H3Ub), as a specific reader for H3K9me3/H3Ub, with the recognition mode distinct from the typical trimethyl-lysine reader. Disruption of the interaction between RFTS and the H3K9me3Ub affects the localization of DNMT1 in stem cells and profoundly impairs the global DNA methylation and genomic stability. Together, this study reveals a previously unappreciated pathway through which H3K9me3 directly reinforces DNMT1-mediated maintenance DNA methylation.
- Department of Biochemistry, University of California, Riverside, CA 92521.
Organizational Affiliation: