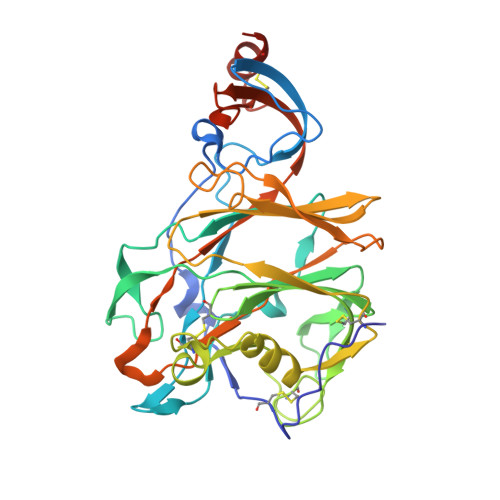
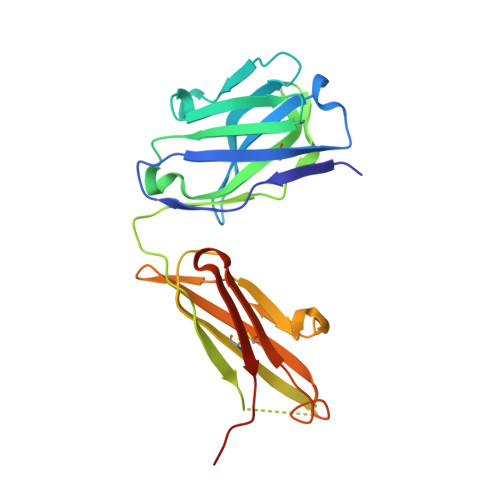
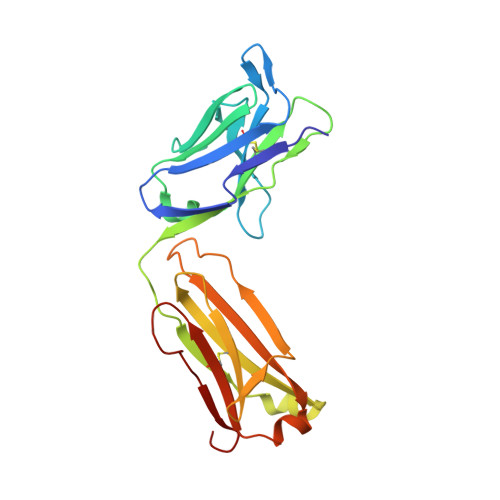
Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Wang, N., Rosen, O., Wang, L., Turner, H.L., Stevens, L.J., Corbett, K.S., Bowman, C.A., Pallesen, J., Shi, W., Zhang, Y., Leung, K., Kirchdoerfer, R.N., Becker, M.M., Denison, M.R., Chappell, J.D., Ward, A.B., Graham, B.S., McLellan, J.S.(2019) Cell Rep 28: 3395-3405.e6
- PubMed: 31553909
- DOI: https://doi.org/10.1016/j.celrep.2019.08.052
- Primary Citation of Related Structures:
6PXG, 6PXH, 6PZ8 - PubMed Abstract:
Middle East respiratory syndrome coronavirus (MERS-CoV) emerged into the human population in 2012 and has caused substantial morbidity and mortality. Potently neutralizing antibodies targeting the receptor-binding domain (RBD) on MERS-CoV spike (S) protein have been characterized, but much less is known about antibodies targeting non-RBD epitopes. Here, we report the structural and functional characterization of G2, a neutralizing antibody targeting the MERS-CoV S1 N-terminal domain (S1-NTD). Structures of G2 alone and in complex with the MERS-CoV S1-NTD define a site of vulnerability comprising two loops, each of which contain a residue mutated in G2-escape variants. Cell-surface binding studies and in vitro competition experiments demonstrate that G2 strongly disrupts the attachment of MERS-CoV S to its receptor, dipeptidyl peptidase-4 (DPP4), with the inhibition requiring the native trimeric S conformation. These results advance our understanding of antibody-mediated neutralization of coronaviruses and should facilitate the development of immunotherapeutics and vaccines against MERS-CoV.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: