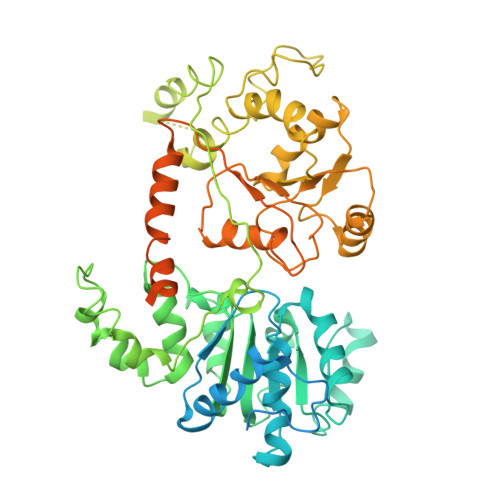
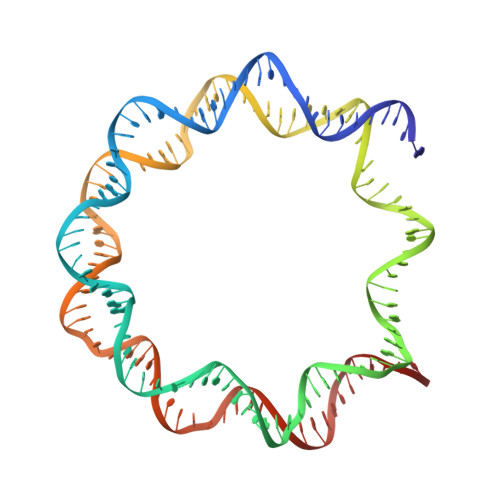
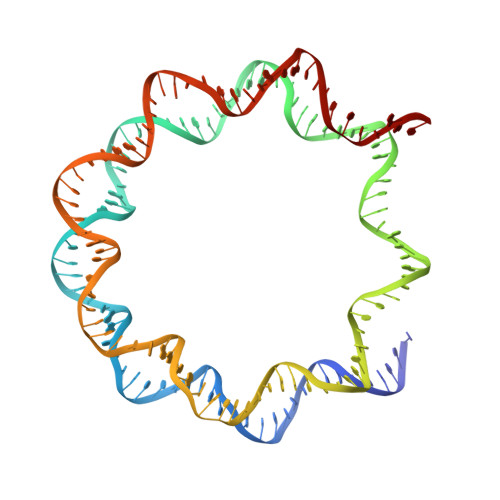
Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome.
Chittori, S., Hong, J., Bai, Y., Subramaniam, S.(2019) Nucleic Acids Res 47: 9400-9409
- PubMed: 31402386
- DOI: https://doi.org/10.1093/nar/gkz670
- Primary Citation of Related Structures:
6PWE, 6PWF - PubMed Abstract:
ATP-dependent chromatin remodeling factors of SWI/SNF2 family including ISWI, SNF2, CHD1 and INO80 subfamilies share a conserved but functionally non-interchangeable ATPase domain. Here we report cryo-electron microscopy (cryo-EM) structures of the nucleosome bound to an ISWI fragment with deletion of the AutoN and HSS regions in nucleotide-free conditions and the free nucleosome at ∼ 4 Å resolution. In the bound conformation, the ATPase domain interacts with the super helical location 2 (SHL 2) of the nucleosomal DNA, with the N-terminal tail of H4 and with the α1 helix of H3. Density for other regions of ISWI is not observed, presumably due to disorder. Comparison with the structure of the free nucleosome reveals that although the histone core remains largely unchanged, remodeler binding causes perturbations in the nucleosomal DNA resulting in a bulge near the SHL2 site. Overall, the structure of the nucleotide-free ISWI-nucleosome complex is similar to the corresponding regions of the recently reported ADP bound ISWI-nucleosome structures, which are significantly different from that observed for the ADP-BeFx bound structure. Our findings are relevant to the initial step of ISWI binding to the nucleosome and provide additional insights into the nucleosome remodeling process driven by ISWI.
- Laboratory of Cell Biology, Center for Cancer Research (CCR), National Cancer Institute (NCI), National Institutes of Health (NIH), Bethesda, MD 20892, USA.
Organizational Affiliation: