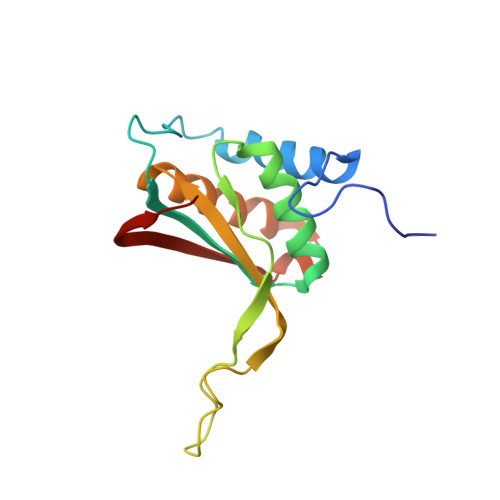
Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.
Malecka, K.A., Dheekollu, J., Deakyne, J.S., Wiedmer, A., Ramirez, U.D., Lieberman, P.M., Messick, T.E.(2019) J Virol 93
- PubMed: 31142669
- DOI: https://doi.org/10.1128/JVI.00487-19
- Primary Citation of Related Structures:
6PW2 - PubMed Abstract:
Epstein-Barr virus is associated with several human malignancies, including nasopharyngeal carcinoma, gastric cancer, and lymphoma. Latently infected cells carry a circularized EBV episome where the origin of replication ( oriP ) is comprised of two elements: the family of repeats (FR) and dyad symmetry (DS). The viral protein Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) binds to FR and DS to promote EBV episome maintenance and DNA replication during latent infection in proliferating cells. EBNA1 binding to the DS constitutes a minimal origin of DNA replication. Here we report the crystal structure of two EBNA1 DNA-binding domain dimers bound to a DS half-site. This structure shows that the DNA is smoothly bent, allowing for stabilizing interactions between the dimers. The dimer-dimer interface requires an intricate hydrogen bonding network involving residues R491 and D581. When this interface is disrupted, we note loss of stable dimer-dimer complex formation on the DNA, compromised oriP -containing plasmid replication in cells, and impaired recruitment of the MCM3 complex to the oriP Surface conservation analysis reveals that these residues are part of a larger conserved surface that may be critical for recruitment of replication machinery to the oriP Our results reveal a new region of EBNA1 critical for its activity and one that may be exploited by targeted small molecules to treat EBV-associated disease. IMPORTANCE Epstein-Barr virus (EBV) is a causative agent of various malignancies and may also contribute to autoimmune disease. The latent and episomal form of the virus is known to drive EBV-associated oncogenesis. Persistence of the viral episome in proliferating tumor cells requires the interaction of Epstein-Barr virus nuclear antigen 1 (EBNA1) with the viral origin of plasmid replication ( oriP ). The dyad symmetry (DS) element in oriP is the essential minimal replicator of oriP Here we report the X-ray crystal structure of EBNA1 bound to DS. The structure reveals a previous unrecognized interface formed between dimers of EBNA1 necessary for cooperative DNA binding, recruitment of cellular replication machinery, and replication function. These findings provide new insights into the mechanism of EBNA1 function at the replication origin and new opportunities to inhibit EBV latent infection and pathogenesis.
- The Wistar Institute, Philadelphia, Pennsylvania, USA.
Organizational Affiliation: