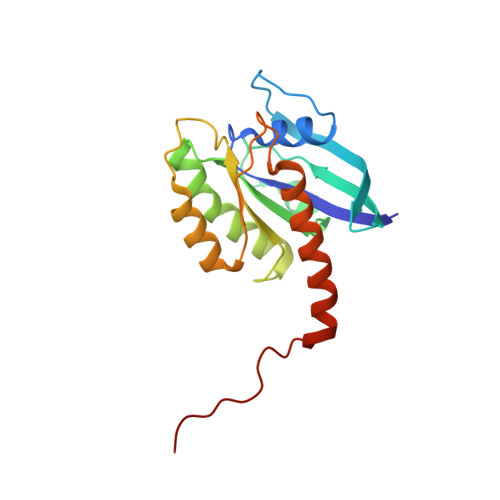
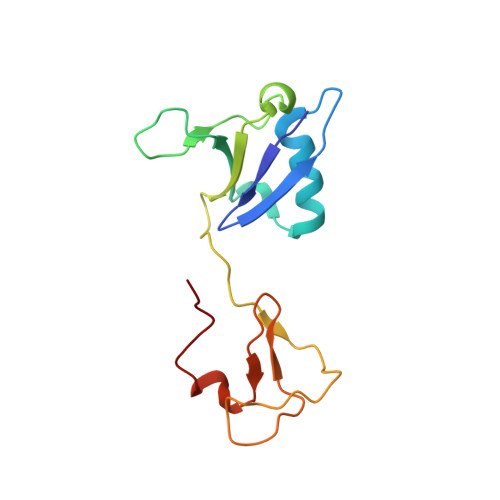
Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Fang, Z., Lee, K.Y., Huo, K.G., Gasmi-Seabrook, G., Zheng, L., Moghal, N., Tsao, M.S., Ikura, M., Marshall, C.B.(2020) Proc Natl Acad Sci U S A 117: 12101-12108
- PubMed: 32414921
- DOI: https://doi.org/10.1073/pnas.1914076117
- Primary Citation of Related Structures:
6PTS, 6PTW - PubMed Abstract:
Membrane anchoring of farnesylated KRAS is critical for activation of RAF kinases, yet our understanding of how these proteins interact on the membrane is limited to isolated domains. The RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF engage KRAS and the plasma membrane, unleashing the kinase domain from autoinhibition. Due to experimental challenges, structural insight into this tripartite KRAS:RBD-CRD:membrane complex has relied on molecular dynamics simulations. Here, we report NMR studies of the KRAS:CRAF RBD-CRD complex. We found that the nucleotide-dependent KRAS-RBD interaction results in transient electrostatic interactions between KRAS and CRD, and we mapped the membrane interfaces of the CRD, RBD-CRD, and the KRAS:RBD-CRD complex. RBD-CRD exhibits dynamic interactions with the membrane through the canonical CRD lipid-binding site (CRD β7-8), as well as an alternative interface comprising β6 and the C terminus of CRD and β2 of RBD. Upon complex formation with KRAS, two distinct states were observed by NMR: State A was stabilized by membrane association of CRD β7-8 and KRAS α4-α5 while state B involved the C terminus of CRD, β3-5 of RBD, and part of KRAS α5. Notably, α4-α5, which has been proposed to mediate KRAS dimerization, is accessible only in state B. A cancer-associated mutation on the state B membrane interface of CRAF RBD (E125K) stabilized state B and enhanced kinase activity and cellular MAPK signaling. These studies revealed a dynamic picture of the assembly of the KRAS-CRAF complex via multivalent and dynamic interactions between KRAS, CRAF RBD-CRD, and the membrane.
- Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 2C4, Canada.
Organizational Affiliation: