Structure of the self-association domain of the chromatin looping factor LDB1
Macindoe, I., Silva, A., Guss, J.M., Mackay, J.P., Matthews, J.M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LIM domain-binding protein 1 | 192 | Mus musculus | Mutation(s): 0 Gene Names: Ldb1, Nli | 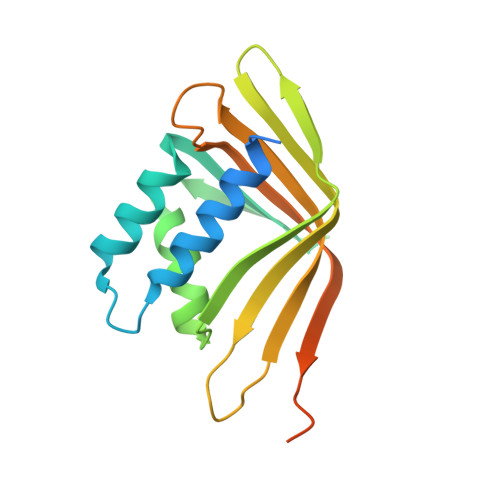 | |
UniProt | |||||
Find proteins for P70662 (Mus musculus) Explore P70662 Go to UniProtKB: P70662 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P70662 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.869 | α = 90 |
| b = 81.869 | β = 90 |
| c = 66.828 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Health and Medical Research Council (NHMRC, Australia) | Australia | APP1064442 |
| Australian Research Council (ARC) | Australia | DP190102543 |