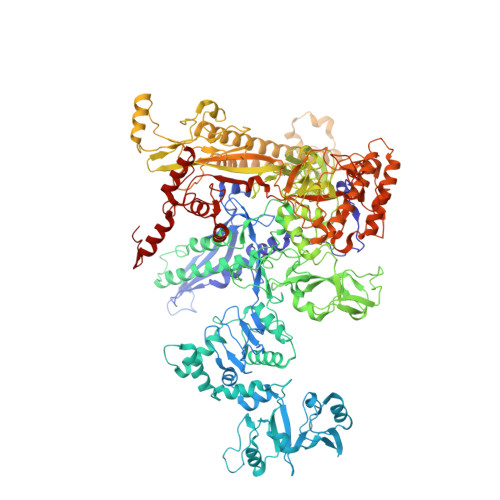
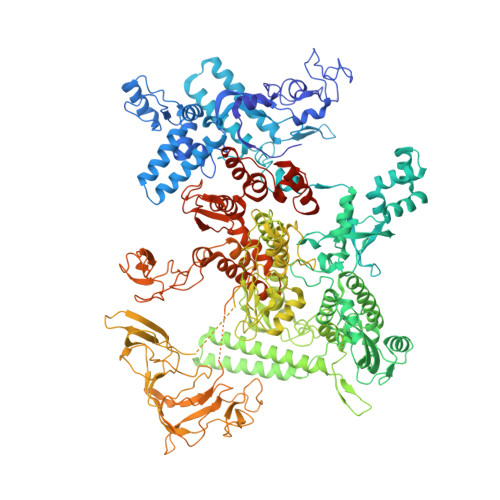
Stepwise Promoter Melting by Bacterial RNA Polymerase.
Chen, J., Chiu, C., Gopalkrishnan, S., Chen, A.Y., Olinares, P.D.B., Saecker, R.M., Winkelman, J.T., Maloney, M.F., Chait, B.T., Ross, W., Gourse, R.L., Campbell, E.A., Darst, S.A.(2020) Mol Cell 78: 275
- PubMed: 32160514
- DOI: https://doi.org/10.1016/j.molcel.2020.02.017
- Primary Citation of Related Structures:
6PSQ, 6PSR, 6PSS, 6PST, 6PSU, 6PSV, 6PSW - PubMed Abstract:
Transcription initiation requires formation of the open promoter complex (RPo). To generate RPo, RNA polymerase (RNAP) unwinds the DNA duplex to form the transcription bubble and loads the DNA into the RNAP active site. RPo formation is a multi-step process with transient intermediates of unknown structure. We use single-particle cryoelectron microscopy to visualize seven intermediates containing Escherichia coli RNAP with the transcription factor TraR en route to forming RPo. The structures span the RPo formation pathway from initial recognition of the duplex promoter in a closed complex to the final RPo. The structures and supporting biochemical data define RNAP and promoter DNA conformational changes that delineate steps on the pathway, including previously undetected transient promoter-RNAP interactions that contribute to populating the intermediates but do not occur in RPo. Our work provides a structural basis for understanding RPo formation and its regulation, a major checkpoint in gene expression throughout evolution.
- Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY 10065, USA.
Organizational Affiliation: