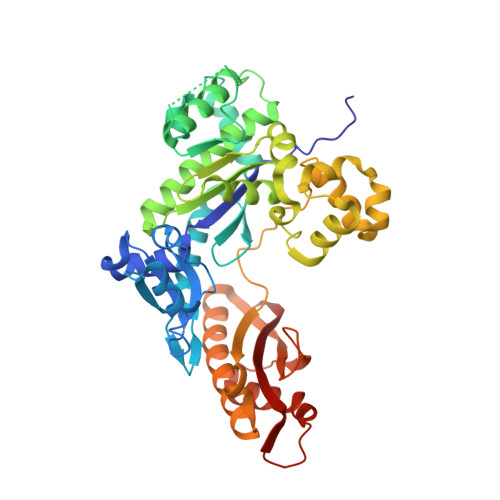
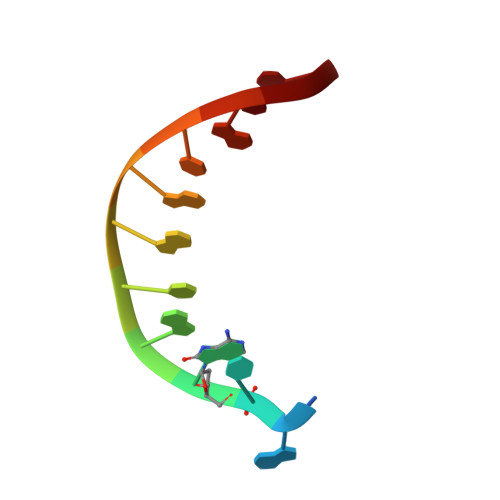
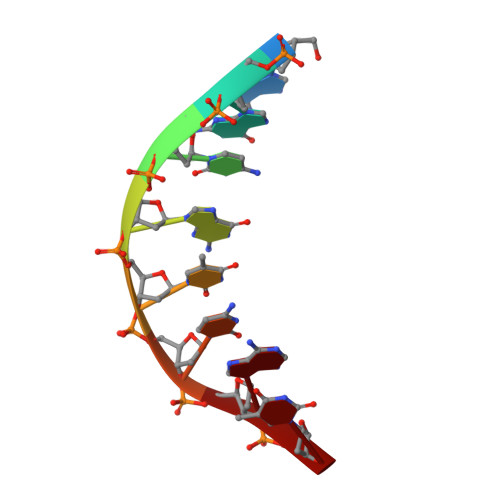
Mutagenesis mechanism of the major oxidative adenine lesion 7,8-dihydro-8-oxoadenine.
Koag, M.C., Jung, H., Lee, S.(2020) Nucleic Acids Res 48: 5119-5134
- PubMed: 32282906
- DOI: https://doi.org/10.1093/nar/gkaa193
- Primary Citation of Related Structures:
6PLC - PubMed Abstract:
Reactive oxygen species generate the genotoxic 8-oxoguanine (oxoG) and 8-oxoadenine (oxoA) as major oxidative lesions. The mutagenicity of oxoG is attributed to the lesion's ability to evade the geometric discrimination of DNA polymerases by adopting Hoogsteen base pairing with adenine in a Watson-Crick-like geometry. Compared with oxoG, the mutagenesis mechanism of oxoA, which preferentially induces A-to-C mutations, is poorly understood. In the absence of protein contacts, oxoA:G forms a wobble conformation, the formation of which is suppressed in the catalytic site of most DNA polymerases. Interestingly, human DNA polymerase η (polη) proficiently incorporates dGTP opposite oxoA, suggesting the nascent oxoA:dGTP overcomes the geometric discrimination of polη. To gain insights into oxoA-mediated mutagenesis, we determined crystal structures of polη bypassing oxoA. When paired with dGTP, oxoA adopted a syn-conformation and formed Hoogsteen pairing while in a wobble geometry, which was stabilized by Gln38-mediated minor groove contacts to oxoA:dGTP. Gln38Ala mutation reduced misinsertion efficiency ∼55-fold, indicating oxoA:dGTP misincorporation was promoted by minor groove interactions. Also, the efficiency of oxoA:dGTP insertion by the X-family polβ decreased ∼380-fold when Asn279-mediated minor groove contact to dGTP was abolished. Overall, these results suggest that, unlike oxoG, oxoA-mediated mutagenesis is greatly induced by minor groove interactions.
- The Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: