Structure of human DNA polymerase beta N279A complexed with 8OA in the template base paired with incoming non-hydrolyzable GTP
Koag, M.C., Lee, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase beta | 335 | Homo sapiens | Mutation(s): 1 Gene Names: POLB EC: 2.7.7.7 (PDB Primary Data), 4.2.99 (PDB Primary Data), 4.2.99.18 (UniProt) | 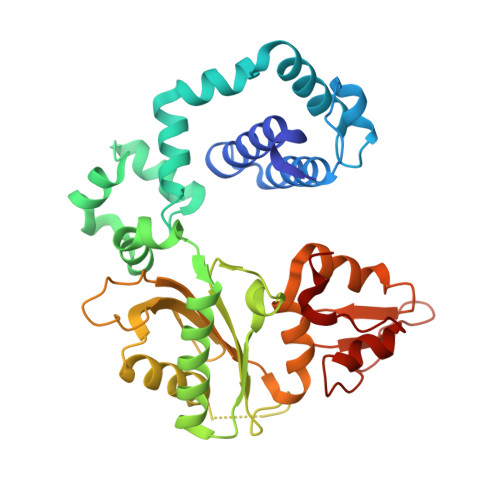 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06746 (Homo sapiens) Explore P06746 Go to UniProtKB: P06746 | |||||
PHAROS: P06746 GTEx: ENSG00000070501 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06746 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*CP*GP*AP*CP*(A38) P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3') | B [auth T] | 16 | synthetic construct | 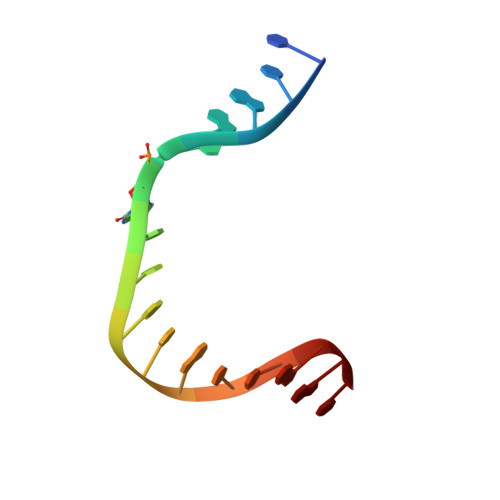 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3') | C [auth P] | 10 | synthetic construct | 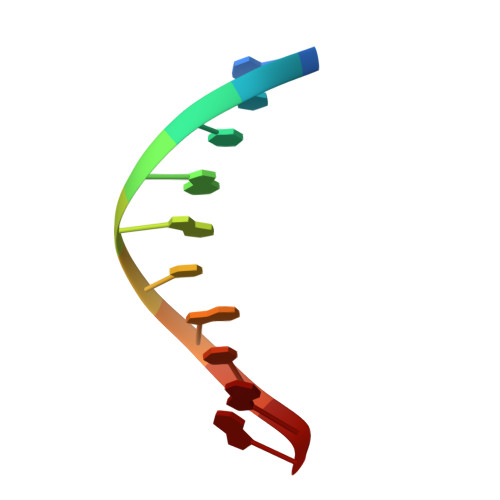 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*GP*TP*CP*GP*G)-3') | 5 | synthetic construct | 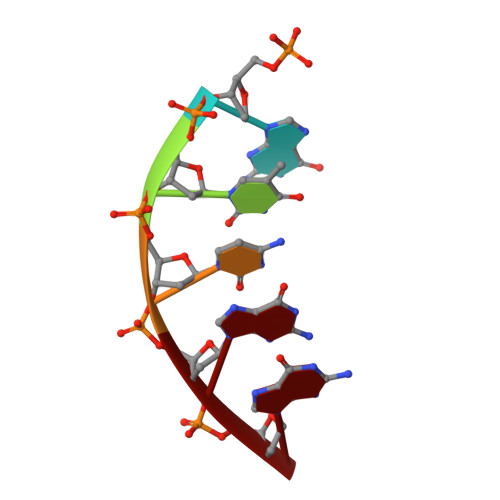 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| XG4 (Subject of Investigation/LOI) Query on XG4 | H [auth A] | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine C10 H17 N6 O12 P3 DWGAAFQEGIMTIA-KVQBGUIXSA-N |  | ||
| MG Query on MG | G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | E [auth A], F [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.609 | α = 90 |
| b = 78.555 | β = 106.83 |
| c = 54.776 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| HKL-2000 | data scaling |
| REFMAC | phasing |
| PHENIX | refinement |