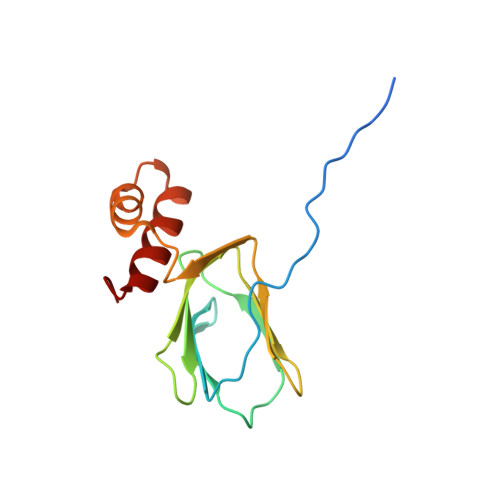
Crystal structure of ATP synthase epsion chain ATP synthase epsilon chain (ATP synthase F1 sector epsilon subunit) (F-ATPase epsilon subunit) from Mycobacterium smegmatis
Abendroth, J., Dranow, D.M., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.