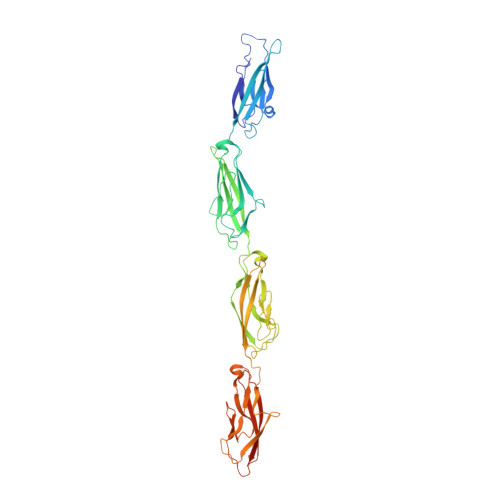A complete Protocadherin-19 ectodomain model for evaluating epilepsy-causing mutations and potential protein interaction sites.
Hudson, J.D., Tamilselvan, E., Sotomayor, M., Cooper, S.R.(2021) Structure 29: 1128-1143.e4
- PubMed: 34520737
- DOI: https://doi.org/10.1016/j.str.2021.07.006
- Primary Citation of Related Structures:
6PGW - PubMed Abstract:
Cadherin superfamily members play a critical role in differential adhesion during neurodevelopment, and their disruption has been linked to several neurodevelopmental disorders. Mutations in protocadherin-19 (PCDH19), a member of the δ-protocadherin subfamily of cadherins, cause a unique form of epilepsy called PCDH19 clustering epilepsy. While PCDH19 and other non-clustered δ-protocadherins form multimers with other members of the cadherin superfamily to alter adhesiveness, the specific protein surfaces responsible for these interactions are unknown. Only portions of the PCDH19 extracellular domain structure had been solved previously. Here, we present a structure of the missing segment from zebrafish Protocadherin-19 (Pcdh19) and create a complete ectodomain model. This model shows the structural environment for 97% of disease-causing missense mutations and reveals two potential surfaces for intermolecular interactions that could modify Pcdh19's adhesive strength and specificity.
- Department of Science and Mathematics, Cedarville University, 251 N. Main Street, Cedarville, OH 45314, USA.
Organizational Affiliation: