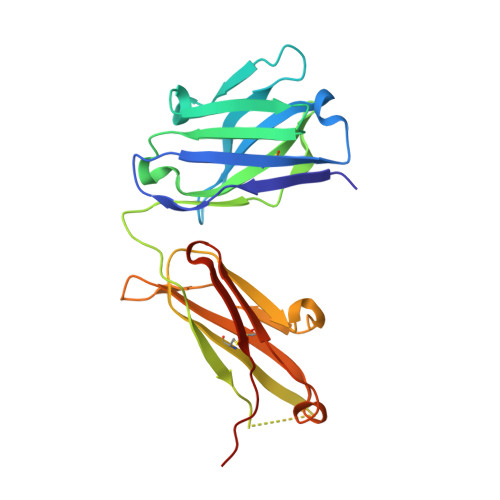
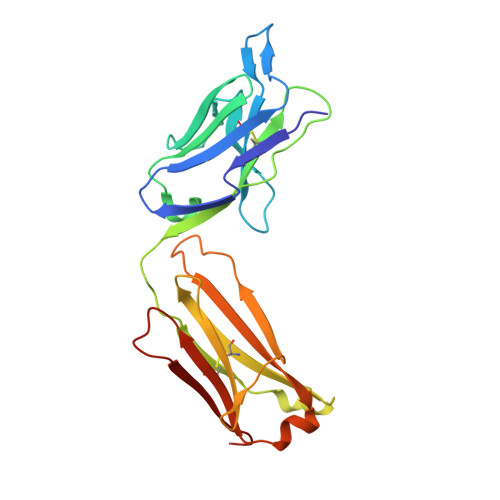
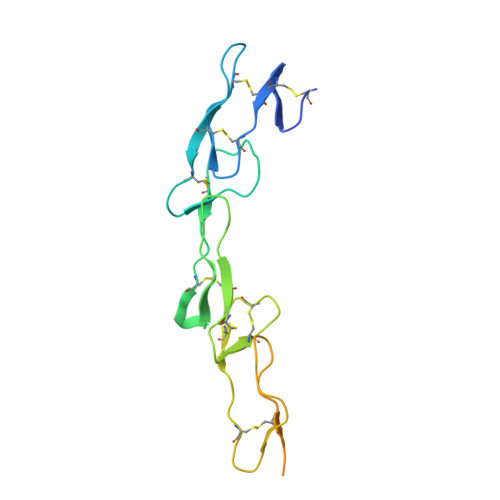
CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
Argiriadi, M.A., Benatuil, L., Dubrovska, I., Egan, D.A., Gao, L., Greischar, A., Hardman, J., Harlan, J., Iyer, R.B., Judge, R.A., Lake, M., Perron, D.C., Sadhukhan, R., Sielaff, B., Sousa, S., Wang, R., McRae, B.L.(2019) BMC Mol Cell Biol 20: 29-29
- PubMed: 31382872
- DOI: https://doi.org/10.1186/s12860-019-0213-4
- Primary Citation of Related Structures:
6PE7, 6PE8, 6PE9 - PubMed Abstract:
CD40 is a 48 kDa type I transmembrane protein that is constitutively expressed on hematopoietic cells such as dendritic cells, macrophages, and B cells. Engagement of CD40 by CD40L expressed on T cells results in the production of proinflammatory cytokines, induces T helper cell function, and promotes macrophage activation. The involvement of CD40 in chronic immune activation has resulted in CD40 being proposed as a therapeutic target for a range of chronic inflammatory diseases. CD40 antagonists are currently being explored for the treatment of autoimmune diseases and several anti-CD40 agonist mAbs have entered clinical development for oncological indications. To better understand the mode of action of anti-CD40 mAbs, we have determined the x-ray crystal structures of the ABBV-323 (anti-CD40 antagonist, ravagalimab) Fab alone, ABBV-323 Fab complexed to human CD40 and FAB516 (anti-CD40 agonist) complexed to human CD40. These three crystals structures 1) identify the conformational CD40 epitope for ABBV-323 recognition 2) illustrate conformational changes which occur in the CDRs of ABBV-323 Fab upon CD40 binding and 3) develop a structural hypothesis for an agonist/antagonist switch in the LCDR1 of this proprietary class of CD40 antibodies. The structure of ABBV-323 Fab demonstrates a unique method for antagonism by stabilizing the proposed functional antiparallel dimer for CD40 receptor via novel contacts to LCDR1, namely residue position R32 which is further supported by a closely related agonist antibody FAB516 which shows only monomeric recognition and no contacts with LCDR1 due to a mutation to L32 on LCDR1. These data provide a structural basis for the full antagonist activity of ABBV-323.
- AbbVie Bioresearch Center, 381 Plantation Street, Worcester, MA, 01605, USA. maria.argiriadi@abbvie.com.
Organizational Affiliation: