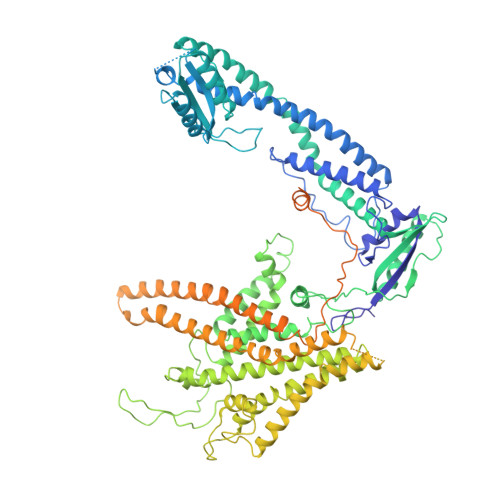
A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Peng, W., Casey, A.K., Fernandez, J., Carpinone, E.M., Servage, K.A., Chen, Z., Li, Y., Tomchick, D.R., Starai, V.J., Orth, K.(2020) Nat Struct Mol Biol 27: 589-597
- PubMed: 32424347
- DOI: https://doi.org/10.1038/s41594-020-0429-1
- Primary Citation of Related Structures:
6PE4, 6PE5 - PubMed Abstract:
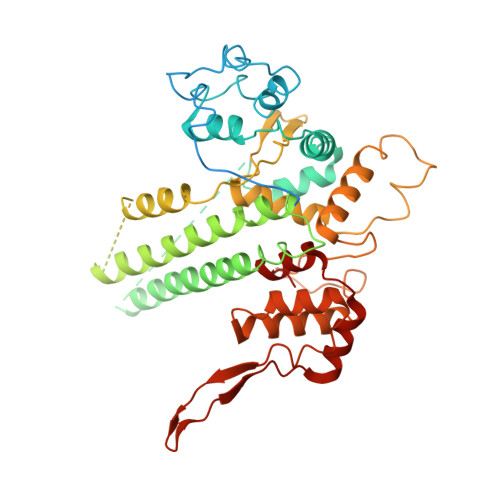
The Vibrio parahaemolyticus T3SS effector VopQ targets host-cell V-ATPase, resulting in blockage of autophagic flux and neutralization of acidic compartments. Here, we report the cryo-EM structure of VopQ bound to the V o subcomplex of the V-ATPase. VopQ inserts into membranes and forms an unconventional pore while binding directly to subunit c of the V-ATPase membrane-embedded subcomplex V o . We show that VopQ arrests yeast growth in vivo by targeting the immature V o subcomplex in the endoplasmic reticulum (ER), thus providing insight into the observation that VopQ kills cells in the absence of a functional V-ATPase. VopQ is a bacterial effector that has been discovered to inhibit a host-membrane megadalton complex by coincidentally binding its target, inserting into a membrane and disrupting membrane potential. Collectively, our results reveal a mechanism by which bacterial effectors modulate host cell biology and provide an invaluable tool for future studies on V-ATPase-mediated membrane fusion and autophagy.
- Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: