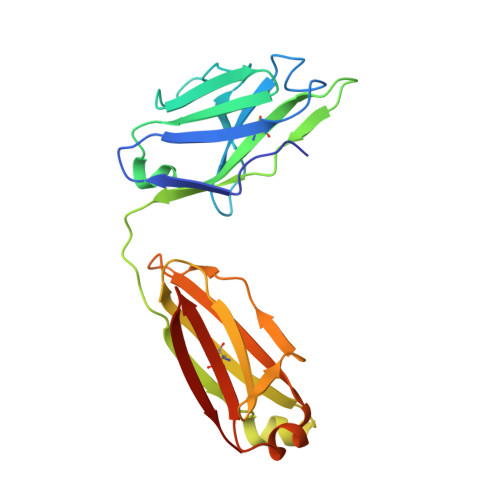
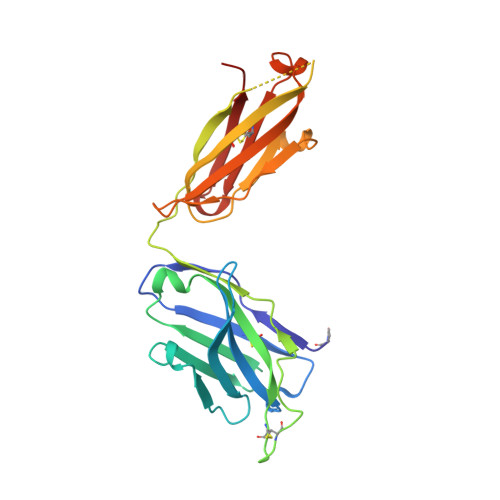
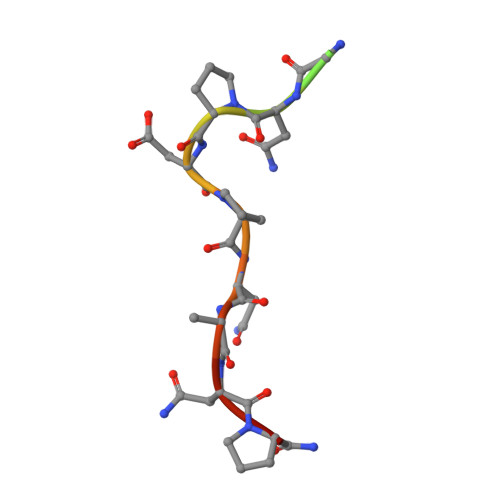
Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Oyen, D., Torres, J.L., Aoto, P.C., Flores-Garcia, Y., Binter, S., Pholcharee, T., Carroll, S., Reponen, S., Wash, R., Liang, Q., Lemiale, F., Locke, E., Bradley, A., King, C.R., Emerling, D., Kellam, P., Zavala, F., Ward, A.B., Wilson, I.A.(2020) PLoS Pathog 16: e1008373-e1008373
- PubMed: 32150583
- DOI: https://doi.org/10.1371/journal.ppat.1008373
- Primary Citation of Related Structures:
6PBV, 6PBW - PubMed Abstract:
Lasting protection has long been a goal for malaria vaccines. The major surface antigen on Plasmodium falciparum sporozoites, the circumsporozoite protein (PfCSP), has been an attractive target for vaccine development and most protective antibodies studied to date interact with the central NANP repeat region of PfCSP. However, it remains unclear what structural and functional characteristics correlate with better protection by one antibody over another. Binding to the junctional region between the N-terminal domain and central NANP repeats has been proposed to result in superior protection: this region initiates with the only NPDP sequence followed immediately by NANP. Here, we isolated antibodies in Kymab mice immunized with full-length recombinant PfCSP and two protective antibodies were selected for further study with reactivity against the junctional region. X-ray and EM structures of two monoclonal antibodies, mAb667 and mAb668, shed light on their differential affinity and specificity for the junctional region. Importantly, these antibodies also bind to the NANP repeat region with equal or better affinity. A comparison with an NANP-only binding antibody (mAb317) revealed roughly similar but statistically distinct levels of protection against sporozoite challenge in mouse liver burden models, suggesting that junctional antibody protection might relate to the ability to also cross-react with the NANP repeat region. Our findings indicate that additional efforts are necessary to isolate a true junctional antibody with no or much reduced affinity to the NANP region to elucidate the role of the junctional epitope in protection.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, United States of America.
Organizational Affiliation: