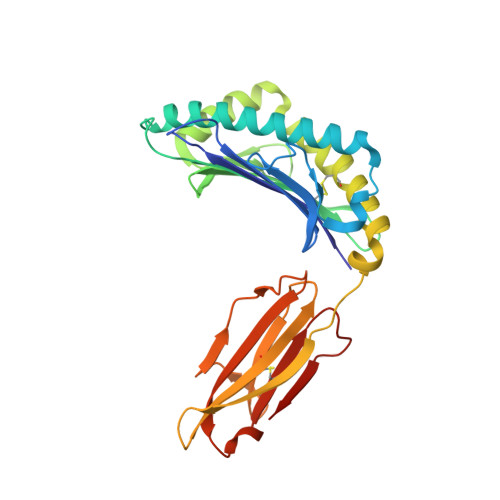
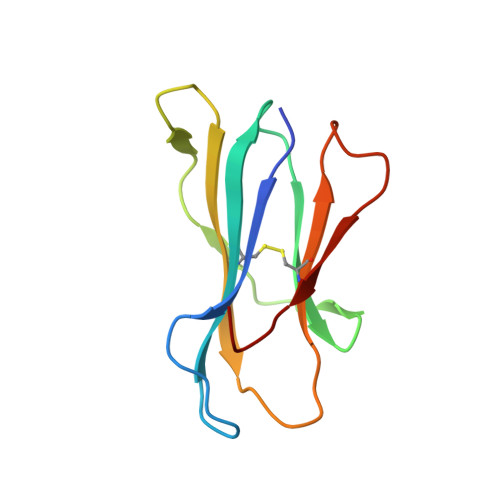
Challenging immunodominance of influenza-specific CD8+T cell responses restricted by the risk-associated HLA-A*68:01 allomorph.
van de Sandt, C.E., Clemens, E.B., Grant, E.J., Rowntree, L.C., Sant, S., Halim, H., Crowe, J., Cheng, A.C., Kotsimbos, T.C., Richards, M., Miller, A., Tong, S.Y.C., Rossjohn, J., Nguyen, T.H.O., Gras, S., Chen, W., Kedzierska, K.(2019) Nat Commun 10: 5579-5579
- PubMed: 31811120
- DOI: https://doi.org/10.1038/s41467-019-13346-4
- Primary Citation of Related Structures:
6PBH - PubMed Abstract:
Although influenza viruses lead to severe illness in high-risk populations, host genetic factors associated with severe disease are largely unknown. As the HLA-A*68:01 allele can be linked to severe pandemic 2009-H1N1 disease, we investigate a potential impairment of HLA-A*68:01-restricted CD8 + T cells to mount robust responses. We elucidate the HLA-A*68:01 + CD8 + T cell response directed toward an extended influenza-derived nucleoprotein (NP) peptide and show that only ~35% individuals have immunodominant A68/NP 145 + CD8 + T cell responses. Dissecting A68/NP 145 + CD8 + T cells in low vs. medium/high responders reveals that high responding donors have A68/NP 145 + CD8 + memory T cells with clonally expanded TCRαβs, while low-responders display A68/NP 145 + CD8 + T cells with predominantly naïve phenotypes and non-expanded TCRαβs. Single-cell index sorting and TCRαβ analyses link expansion of A68/NP 145 + CD8 + T cells to their memory potential. Our study demonstrates the immunodominance potential of influenza-specific CD8 + T cells presented by a risk HLA-A*68:01 molecule and advocates for priming CD8 + T cell compartments in HLA-A*68:01-expressing individuals for establishment of pre-existing protective memory T cell pools.
- Department of Microbiology and Immunology, University of Melbourne at The Peter Doherty Institute, Melbourne, VIC, 3000, Australia.
Organizational Affiliation: