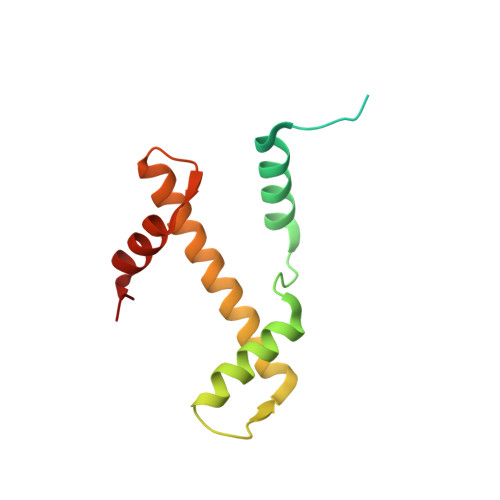
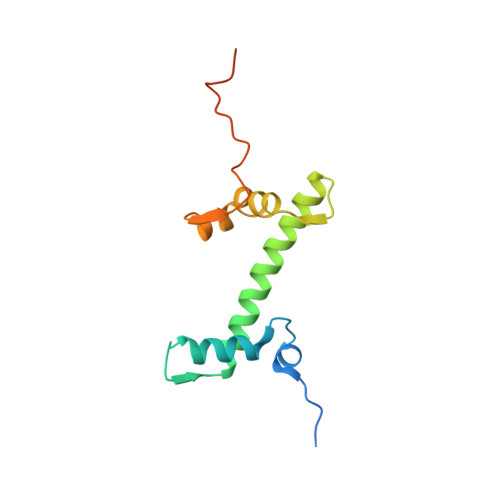
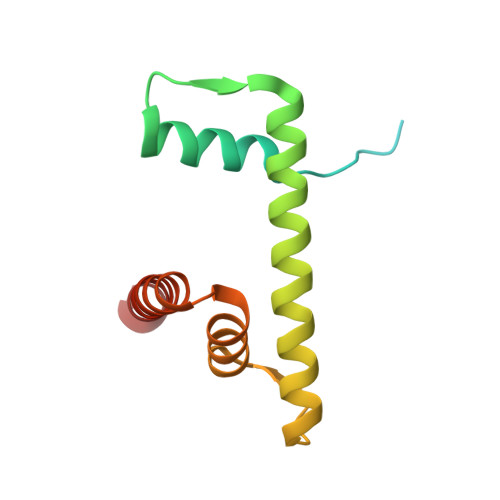
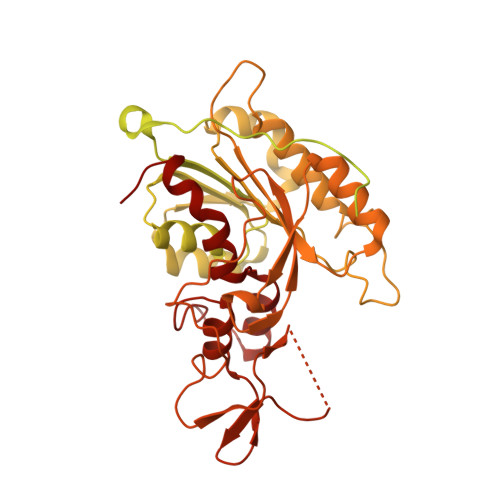
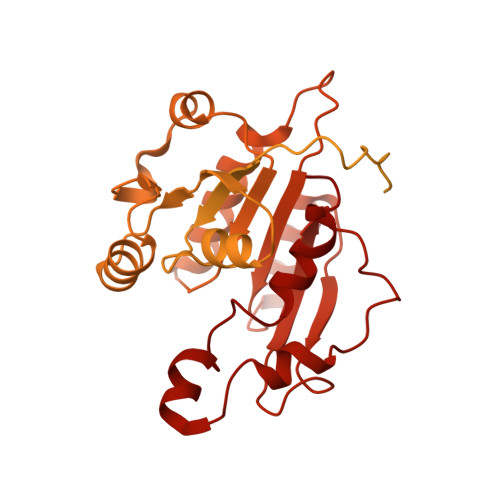
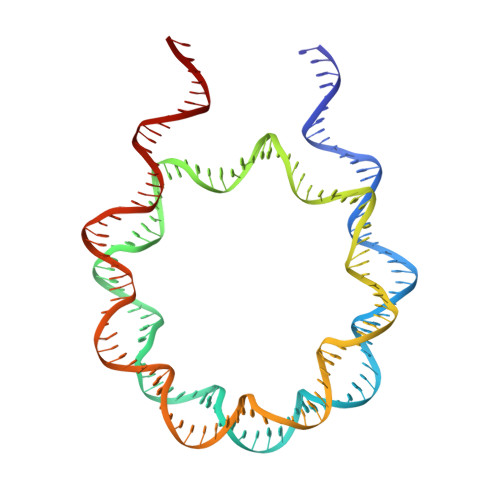
Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Xu, T.H., Liu, M., Zhou, X.E., Liang, G., Zhao, G., Xu, H.E., Melcher, K., Jones, P.A.(2020) Nature 586: 151-155
- PubMed: 32968275
- DOI: https://doi.org/10.1038/s41586-020-2747-1
- Primary Citation of Related Structures:
6PA7 - PubMed Abstract:
CpG methylation by de novo DNA methyltransferases (DNMTs) 3A and 3B is essential for mammalian development and differentiation and is frequently dysregulated in cancer 1 . These two DNMTs preferentially bind to nucleosomes, yet cannot methylate the DNA wrapped around the nucleosome core 2 , and they favour the methylation of linker DNA at positioned nucleosomes 3,4 . Here we present the cryo-electron microscopy structure of a ternary complex of catalytically competent DNMT3A2, the catalytically inactive accessory subunit DNMT3B3 and a nucleosome core particle flanked by linker DNA. The catalytic-like domain of the accessory DNMT3B3 binds to the acidic patch of the nucleosome core, which orients the binding of DNMT3A2 to the linker DNA. The steric constraints of this arrangement suggest that nucleosomal DNA must be moved relative to the nucleosome core for de novo methylation to occur.
- Center for Cancer and Cell Biology, Program for Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.
Organizational Affiliation: