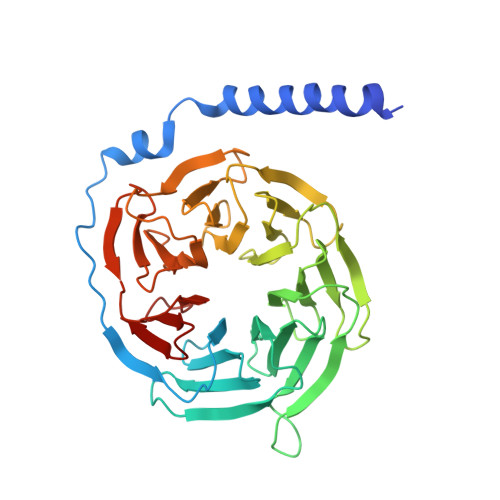
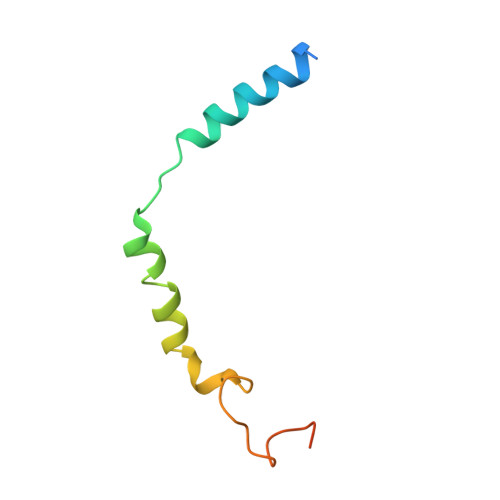
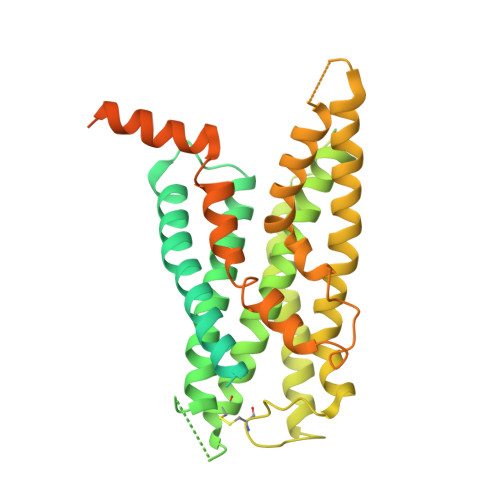
Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Liang, Y.L., Belousoff, M.J., Zhao, P., Koole, C., Fletcher, M.M., Truong, T.T., Julita, V., Christopoulos, G., Xu, H.E., Zhang, Y., Khoshouei, M., Christopoulos, A., Danev, R., Sexton, P.M., Wootten, D.(2020) Mol Cell 77: 656-668.e5
- PubMed: 32004469
- DOI: https://doi.org/10.1016/j.molcel.2020.01.012
- Primary Citation of Related Structures:
6P9X, 6P9Y - PubMed Abstract:
Class B G protein-coupled receptors (GPCRs) are important therapeutic targets for major diseases. Here, we present structures of peptide and Gs-bound pituitary adenylate cyclase-activating peptide, PAC1 receptor, and corticotropin-releasing factor (CRF), (CRF1) receptor. Together with recently solved structures, these provide coverage of the major class B GPCR subfamilies. Diverse orientations of the extracellular domain to the receptor core in different receptors are at least partially dependent on evolutionary conservation in the structure and nature of peptide interactions. Differences in peptide interactions to the receptor core also influence the interlinked TM2-TM1-TM6/ECL3/TM7 domain, and this is likely important in their diverse signaling. However, common conformational reorganization of ECL2, linked to reorganization of ICL2, modulates G protein contacts. Comparison between receptors reveals ICL2 as a key domain forming dynamic G protein interactions in a receptor- and ligand-specific manner. This work advances our understanding of class B GPCR activation and Gs coupling.
- Drug Discovery Biology and Department of Pharmacology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, VIC, Australia.
Organizational Affiliation: