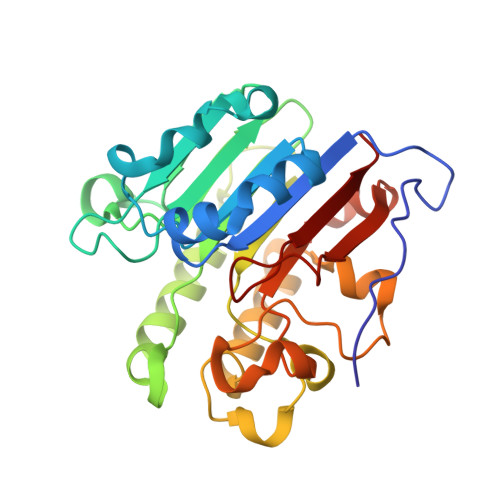
Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
McNeill, D.R., Whitaker, A.M., Stark, W.J., Illuzzi, J.L., McKinnon, P.J., Freudenthal, B.D., Wilson, D.M.(2020) Mutagenesis 35: 27-38
- PubMed: 31816044
- DOI: https://doi.org/10.1093/mutage/gez046
- Primary Citation of Related Structures:
6P93, 6P94 - PubMed Abstract:
DNA is susceptible to a range of chemical modifications, with one of the most frequent lesions being apurinic/apyrimidinic (AP) sites. AP sites arise due to damage-induced (e.g. alkylation) or spontaneous hydrolysis of the N-glycosidic bond that links the base to the sugar moiety of the phosphodiester backbone, or through the enzymatic activity of DNA glycosylases, which release inappropriate bases as part of the base excision repair (BER) response. Unrepaired AP sites, which lack instructional information, have the potential to cause mutagenesis or to arrest progressing DNA or RNA polymerases, potentially causing outcomes such as cellular transformation, senescence or death. The predominant enzyme in humans responsible for repairing AP lesions is AP endonuclease 1 (APE1). Besides being a powerful AP endonuclease, APE1 possesses additional DNA repair activities, such as 3'-5' exonuclease, 3'-phophodiesterase and nucleotide incision repair. In addition, APE1 has been shown to stimulate the DNA-binding activity of a number of transcription factors through its 'REF1' function, thereby regulating gene expression. In this article, we review the structural and biochemical features of this multifunctional protein, while reporting on new structures of the APE1 variants Cys65Ala and Lys98Ala. Using a functional complementation approach, we also describe the importance of the repair and REF1 activities in promoting cell survival, including the proposed passing-the-baton coordination in BER. Finally, results are presented indicating a critical role for APE1 nuclease activities in resistance to the genotoxins methyl methanesulphonate and bleomycin, supporting biologically important functions as an AP endonuclease and 3'-phosphodiesterase, respectively.
- Laboratory of Molecular Gerontology, National Institute on Aging, Intramural Research Program, National Institutes of Health, Baltimore, MD, USA.
Organizational Affiliation: