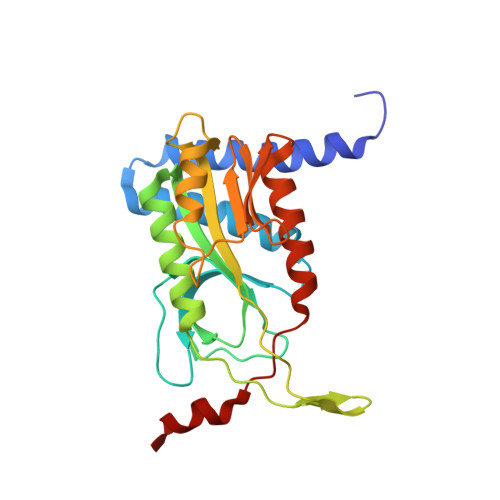
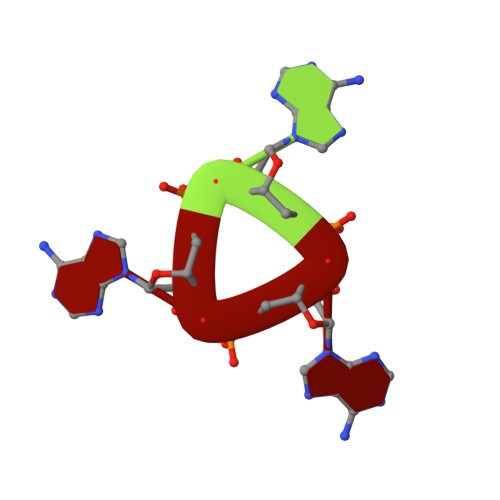
Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Lau, R.K., Ye, Q., Birkholz, E.A., Berg, K.R., Patel, L., Mathews, I.T., Watrous, J.D., Ego, K., Whiteley, A.T., Lowey, B., Mekalanos, J.J., Kranzusch, P.J., Jain, M., Pogliano, J., Corbett, K.D.(2020) Mol Cell 77: 723
- PubMed: 31932164
- DOI: https://doi.org/10.1016/j.molcel.2019.12.010
- Primary Citation of Related Structures:
6P7O, 6P7P, 6P7Q, 6Q1H, 6UXF, 6UXG - PubMed Abstract:
Bacteria possess an array of defenses against foreign invaders, including a broadly distributed bacteriophage defense system termed CBASS (cyclic oligonucleotide-based anti-phage signaling system). In CBASS systems, a cGAS/DncV-like nucleotidyltransferase synthesizes cyclic di- or tri-nucleotide second messengers in response to infection, and these molecules activate diverse effectors to mediate bacteriophage immunity via abortive infection. Here, we show that the CBASS effector NucC is related to restriction enzymes but uniquely assembles into a homotrimer. Binding of NucC trimers to a cyclic tri-adenylate second messenger promotes assembly of a NucC homohexamer competent for non-specific double-strand DNA cleavage. In infected cells, NucC activation leads to complete destruction of the bacterial chromosome, causing cell death prior to completion of phage replication. In addition to CBASS systems, we identify NucC homologs in over 30 type III CRISPR/Cas systems, where they likely function as accessory nucleases activated by cyclic oligoadenylate second messengers synthesized by these systems' effector complexes.
- Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA, USA; Biomedical Sciences Graduate Program, University of California, San Diego, La Jolla, CA, USA.
Organizational Affiliation: