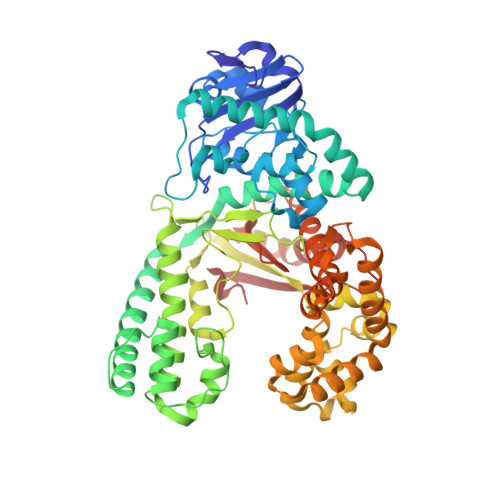
The importance of Ile716 toward the mutagenicity of 8-Oxo-2'-deoxyguanosine with Bacillus fragment DNA polymerase.
Hamm, M.L., Garcia, A.A., Gilbert, R., Johri, M., Ricart, M., Sholes, S.L., Murray-Nerger, L.A., Wu, E.Y.(2020) DNA Repair (Amst) 89: 102826-102826
- PubMed: 32113909
- DOI: https://doi.org/10.1016/j.dnarep.2020.102826
- Primary Citation of Related Structures:
6P5C - PubMed Abstract:
8-oxo-2'-deoxyguanosine (OdG) is a prominent DNA lesion that can direct the incorporation of dCTP or dATP during replication. As the latter reaction can lead to mutation, the ratio of dCTP/dATP incorporation can significantly affect the mutagenic potential of OdG. Previous work with the A-family polymerase BF and seven analogues of OdG identified a major groove amino acid, Ile716, which likely influences the dCTP/dATP incorporation ratio opposite OdG. To further probe the importance of this amino acid, dCTP and dATP incorporations opposite the same seven analogues were tested with two BF mutants, I716M and I716A. Results from these studies support the presence of clashing interactions between Ile716 and the C8-oxygen and C2-amine during dCTP and dATP incorporations, respectively. Crystallographic analysis suggests that residue 716 alters the conformation of the template base prior to insertion into the active site, thereby affecting enzymatic efficiency. These results are also consistent with previous work with A-family polymerases, which indicate they have tight, rigid active sites that are sensitive to template perturbations.
- Department of Chemistry, University of Richmond, 138 UR Drive, Richmond, VA, 23173, United States. Electronic address: mhamm@richmond.edu.
Organizational Affiliation: