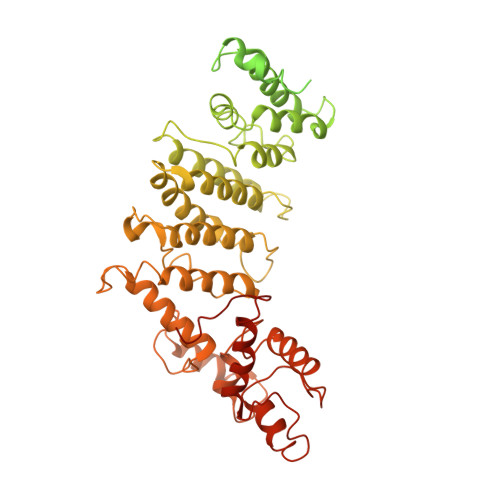
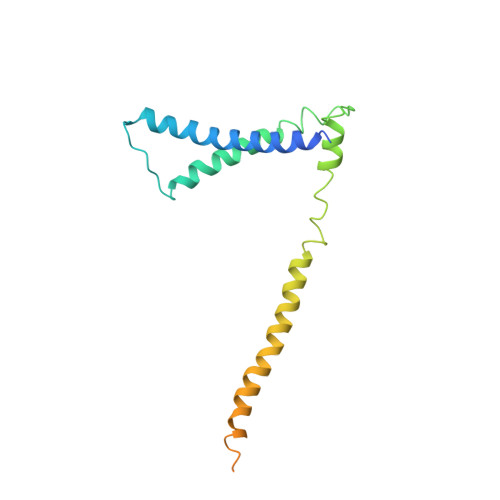
The structure of the yeast Ctf3 complex.
Hinshaw, S.M., Dates, A.N., Harrison, S.C.(2019) Elife 8
- PubMed: 31194673
- DOI: https://doi.org/10.7554/eLife.48215
- Primary Citation of Related Structures:
6OUA - PubMed Abstract:
Kinetochores are the chromosomal attachment points for spindle microtubules. They are also signaling hubs that control major cell cycle transitions and coordinate chromosome folding. Most well-studied eukaryotes rely on a conserved set of factors, which are divided among two loosely-defined groups, for these functions. Outer kinetochore proteins contact microtubules or regulate this contact directly. Inner kinetochore proteins designate the kinetochore assembly site by recognizing a specialized nucleosome containing the H3 variant Cse4/CENP-A. We previously determined the structure, resolved by cryo-electron microscopy (cryo-EM), of the yeast Ctf19 complex (Ctf19c, homologous to the vertebrate CCAN), providing a high-resolution view of inner kinetochore architecture (Hinshaw and Harrison, 2019). We now extend these observations by reporting a near-atomic model of the Ctf3 complex, the outermost Ctf19c sub-assembly seen in our original cryo-EM density. The model is sufficiently well-determined by the new data to enable molecular interpretation of Ctf3 recruitment and function.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Howard Hughes Medical Institute, Boston, United States.
Organizational Affiliation: