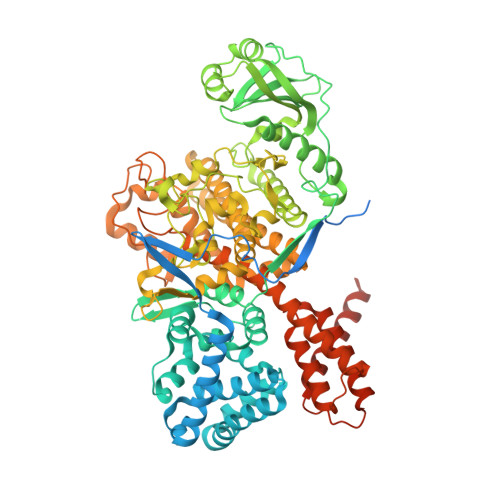
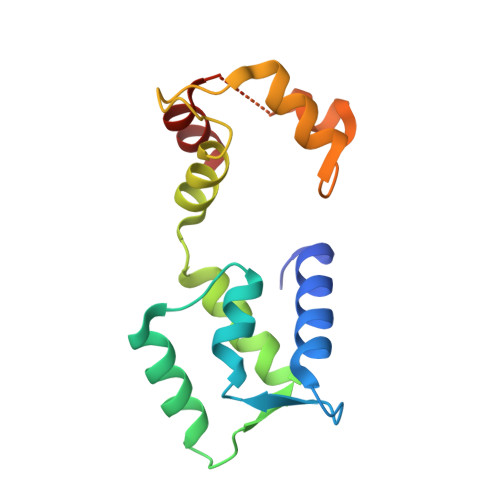
Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases.
Black, M.H., Osinski, A., Gradowski, M., Servage, K.A., Pawlowski, K., Tomchick, D.R., Tagliabracci, V.S.(2019) Science 364: 787-792
- PubMed: 31123136
- DOI: https://doi.org/10.1126/science.aaw7446
- Primary Citation of Related Structures:
6OQQ - PubMed Abstract:
Enzymes with a protein kinase fold transfer phosphate from adenosine 5'-triphosphate (ATP) to substrates in a process known as phosphorylation. Here, we show that the Legionella meta-effector SidJ adopts a protein kinase fold, yet unexpectedly catalyzes protein polyglutamylation. SidJ is activated by host-cell calmodulin to polyglutamylate the SidE family of ubiquitin (Ub) ligases. Crystal structures of the SidJ-calmodulin complex reveal a protein kinase fold that catalyzes ATP-dependent isopeptide bond formation between the amino group of free glutamate and the γ-carboxyl group of an active-site glutamate in SidE. We show that SidJ polyglutamylation of SidE, and the consequent inactivation of Ub ligase activity, is required for successful Legionella replication in a viable eukaryotic host cell.
- Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: