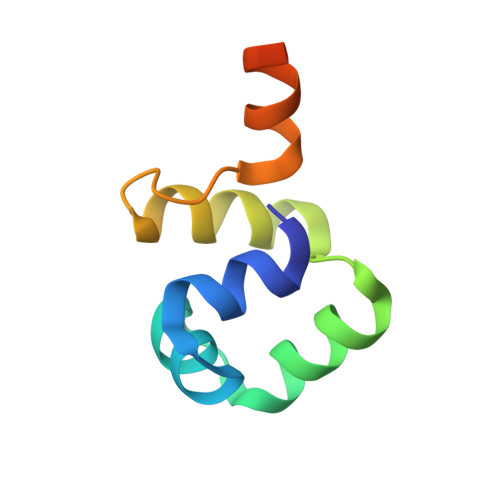
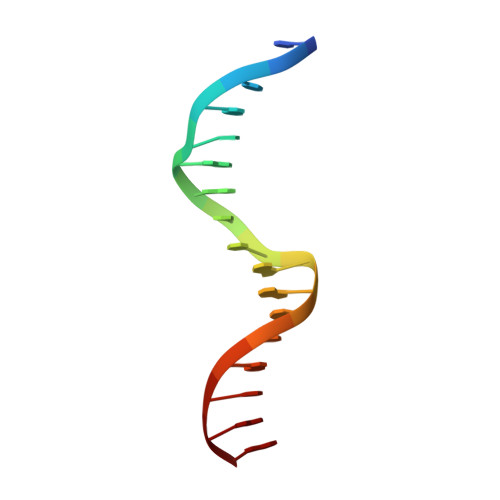
Extreme divergence between one-to-one orthologs: the structure of N15 Cro bound to operator DNA and its relationship to the lambda Cro complex.
Hall, B.M., Roberts, S.A., Cordes, M.H.J.(2019) Nucleic Acids Res 47: 7118-7129
- PubMed: 31180482
- DOI: https://doi.org/10.1093/nar/gkz507
- Primary Citation of Related Structures:
6ON0 - PubMed Abstract:
The gene cro promotes lytic growth of phages through binding of Cro protein dimers to regulatory DNA sites. Most Cro proteins are one-to-one orthologs, yet their sequence, structure and binding site sequences are quite divergent across lambdoid phages. We report the cocrystal structure of bacteriophage N15 Cro with a symmetric consensus site. We contrast this complex with an orthologous structure from phage λ, which has a dissimilar binding site sequence and a Cro protein that is highly divergent in sequence, dimerization interface and protein fold. The N15 Cro complex has less DNA bending and smaller DNA-induced changes in protein structure. N15 Cro makes fewer direct contacts and hydrogen bonds to bases, relying mostly on water-mediated and Van der Waals contacts to recognize the sequence. The recognition helices of N15 Cro and λ Cro make mostly nonhomologous and nonanalogous contacts. Interface alignment scores show that half-site binding geometries of N15 Cro and λ Cro are less similar to each other than to distantly related CI repressors. Despite this divergence, the Cro family shows several code-like protein-DNA sequence covariations. In some cases, orthologous genes can achieve a similar biological function using very different specific molecular interactions.
- Department of Chemistry and Biochemistry, University of Arizona, Tucson, AZ 85721, USA.
Organizational Affiliation: