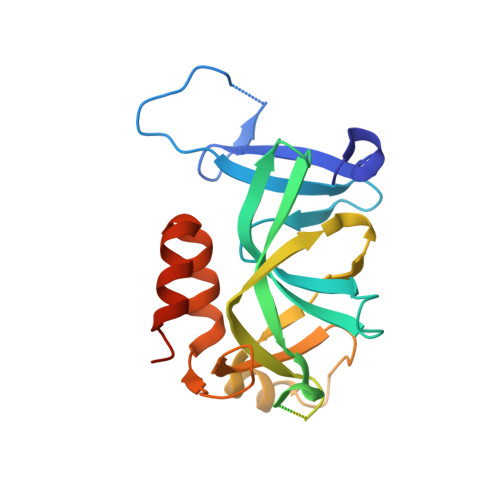
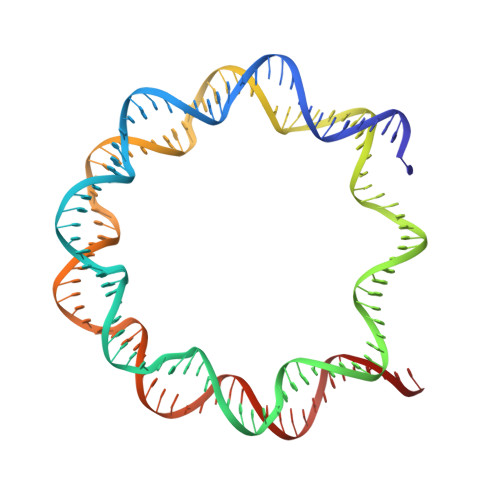
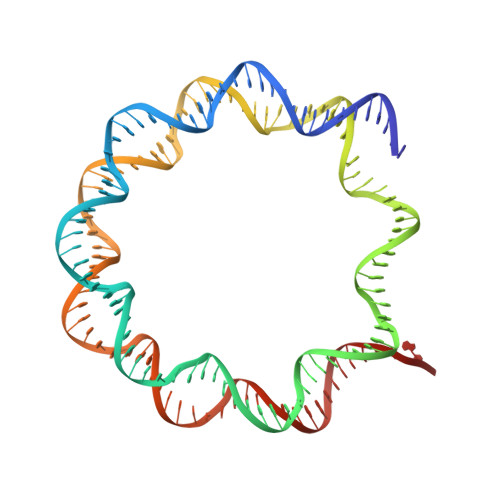
Structure and function of the Orc1 BAH-nucleosome complex.
De Ioannes, P., Leon, V.A., Kuang, Z., Wang, M., Boeke, J.D., Hochwagen, A., Armache, K.J.(2019) Nat Commun 10: 2894-2894
- PubMed: 31263106
- DOI: https://doi.org/10.1038/s41467-019-10609-y
- Primary Citation of Related Structures:
6OM3 - PubMed Abstract:
The Origin Recognition Complex (ORC) is essential for replication, heterochromatin formation, telomere maintenance and genome stability in eukaryotes. Here we present the structure of the yeast Orc1 BAH domain bound to the nucleosome core particle. Our data reveal that Orc1, unlike its close homolog Sir3 involved in gene silencing, does not appear to discriminate between acetylated and non-acetylated lysine 16, modification states of the histone H4 tail that specify open and closed chromatin respectively. We elucidate the mechanism for this unique feature of Orc1 and hypothesize that its ability to interact with nucleosomes regardless of K16 modification state enables it to perform critical functions in both hetero- and euchromatin. We also show that direct interactions with nucleosomes are essential for Orc1 to maintain the integrity of rDNA borders during meiosis, a process distinct and independent from its known roles in silencing and replication.
- Skirball Institute of Biomolecular Medicine, Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, NY, 10016, USA.
Organizational Affiliation: