Structural Intermediates of the Dimeric Kinesin Stepping Cycle Revealed by Cryo-EM
Cha, H.K., Debs, G., Liu, X., Liu, D., Sindelar, C.V.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin beta-2B chain | A [auth B] | 426 | Bos taurus | Mutation(s): 0 | 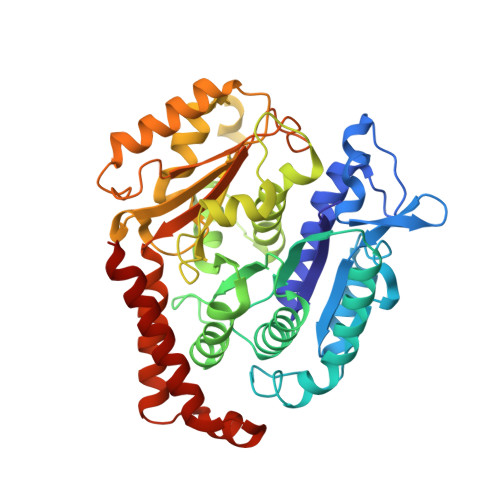 |
UniProt | |||||
Find proteins for Q6B856 (Bos taurus) Explore Q6B856 Go to UniProtKB: Q6B856 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6B856 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin alpha-1B chain | B [auth A] | 437 | Bos taurus | Mutation(s): 0 EC: 3.6.5 | 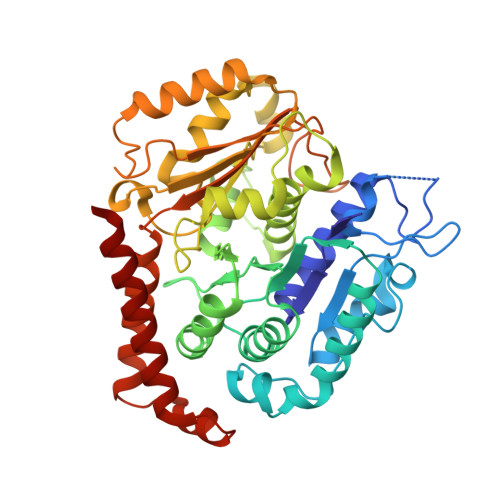 |
UniProt | |||||
Find proteins for P81947 (Bos taurus) Explore P81947 Go to UniProtKB: P81947 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P81947 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Kinesin-1 heavy chain | C [auth K] | 317 | Homo sapiens | Mutation(s): 0 Gene Names: KIF5B, KNS, KNS1 | 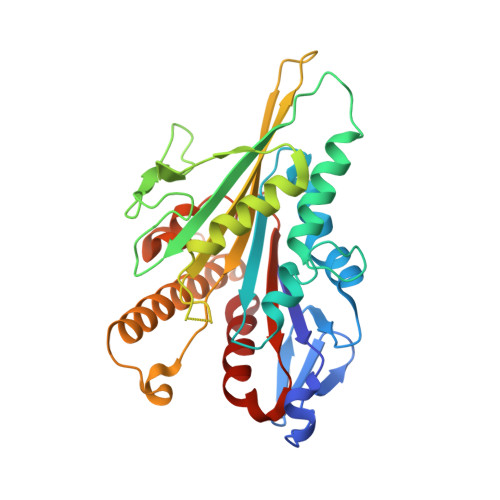 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P33176 (Homo sapiens) Explore P33176 Go to UniProtKB: P33176 | |||||
PHAROS: P33176 GTEx: ENSG00000170759 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P33176 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | F [auth A] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| G2P Query on G2P | D [auth B] | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER C11 H18 N5 O13 P3 GXTIEXDFEKYVGY-KQYNXXCUSA-N |  | ||
| MG Query on MG | E [auth B], G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) | United States | R01 GM 110530-01 |