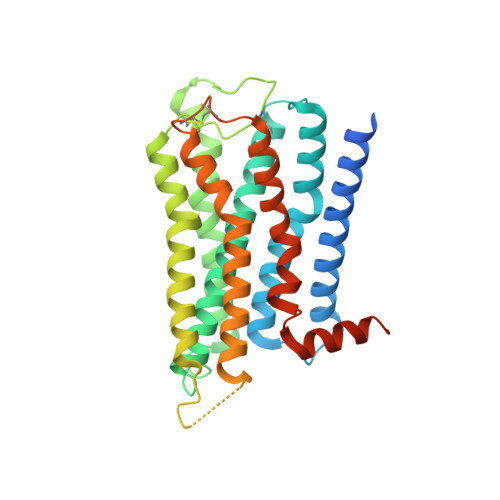
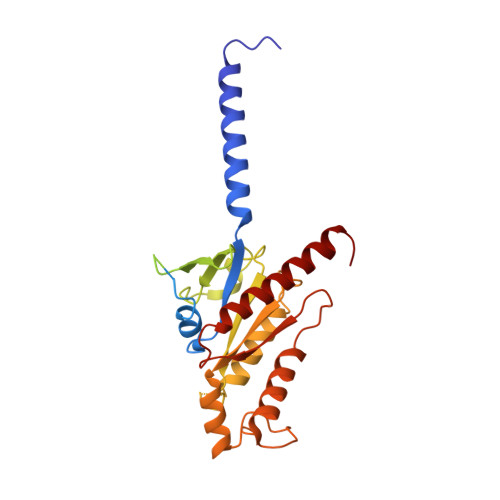
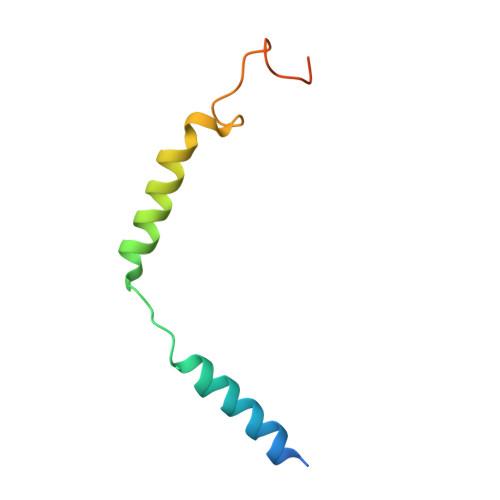
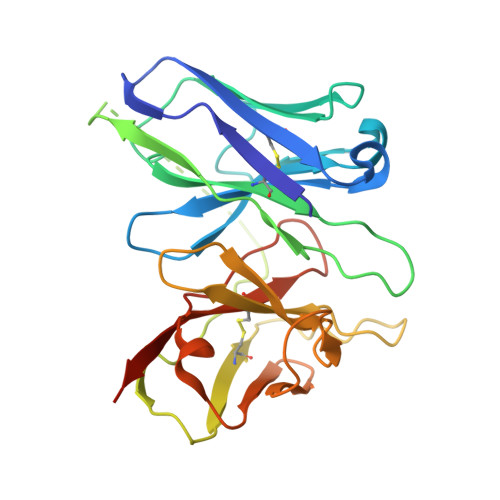
Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes.
Maeda, S., Qu, Q., Robertson, M.J., Skiniotis, G., Kobilka, B.K.(2019) Science 364: 552-557
- PubMed: 31073061
- DOI: https://doi.org/10.1126/science.aaw5188
- Primary Citation of Related Structures:
6OIJ, 6OIK - PubMed Abstract:
Muscarinic acetylcholine receptors are G protein-coupled receptors that respond to acetylcholine and play important signaling roles in the nervous system. There are five muscarinic receptor subtypes (M1R to M5R), which, despite sharing a high degree of sequence identity in the transmembrane region, couple to different heterotrimeric GTP-binding proteins (G proteins) to transmit signals. M1R, M3R, and M5R couple to the G q/ 11 family, whereas M2R and M4R couple to the G i/ o family. Here, we present and compare the cryo-electron microscopy structures of M1R in complex with G 11 and M2R in complex with G oA The M1R-G 11 complex exhibits distinct features, including an extended transmembrane helix 5 and carboxyl-terminal receptor tail that interacts with G protein. Detailed analysis of these structures provides a framework for understanding the molecular determinants of G-protein coupling selectivity.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.
Organizational Affiliation: