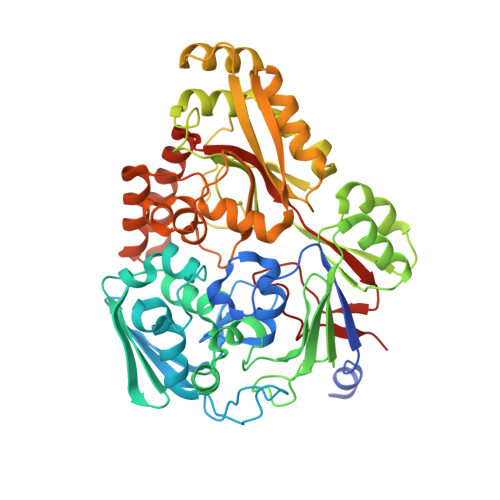
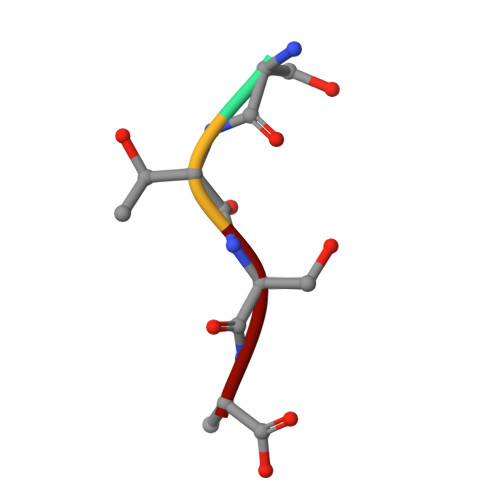
Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
Rahman, M.M., Machuca, M.A., Khan, M.F., Barlow, C.K., Schittenhelm, R.B., Roujeinikova, A.(2019) J Bacteriol 201
- PubMed: 31358613
- DOI: https://doi.org/10.1128/JB.00400-19
- Primary Citation of Related Structures:
6OFQ, 6PU3 - PubMed Abstract:
The gastric pathogen Helicobacter pylori has limited ability to use carbohydrates as a carbon source, relying instead on exogenous amino acids and peptides. Uptake of certain peptides by H. pylori requires an ATP binding cassette (ABC) transporter annotated dipeptide permease (Dpp). The transporter specificity is determined by its cognate substrate-binding protein DppA, which captures ligands in the periplasm and delivers them to the permease. Here, we show that, unlike previously characterized DppA proteins, H. pylori DppA binds, with micromolar affinity, peptides of diverse amino acid sequences ranging between two and eight residues in length. We present analysis of the 1.45-Å-resolution crystal structure of its complex with the tetrapeptide STSA, which provides a structural rationale for the observed broad specificity. Analysis of the molecular surface revealed a ligand-binding pocket that is large enough to accommodate peptides of up to nine residues in length. The structure suggests that H. pylori DppA is able to recognize a wide range of peptide sequences by forming interactions primarily with the peptide main chain atoms. The loop that terminates the peptide-binding pocket in DppAs from other bacteria is significantly shorter in the H. pylori protein, providing an explanation for its ability to bind longer peptides. The subsites accommodating the two N-terminal residues of the peptide ligand make the greatest contribution to the protein-ligand binding energy, in agreement with the observation that dipeptides bind with affinity close to that of longer peptides. IMPORTANCE The World Health Organization listed Helicobacter pylori as a high-priority pathogen for antibiotic development. The potential of using peptide transporters in drug design is well recognized. We discovered that the substrate-binding protein of the ABC transporter for peptides, termed dipeptide permease, is an unusual member of its family in that it directly binds peptides of diverse amino acid sequences, ranging between two and eight residues in length. We also provided a structural rationale for the observed broad specificity. Since the ability to import peptides as a source of carbon is critical for H. pylori , our findings will inform drug design strategies based on inhibition or fusion of membrane-impermeant antimicrobials with peptides.
- Infection and Immunity Program, Monash Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: