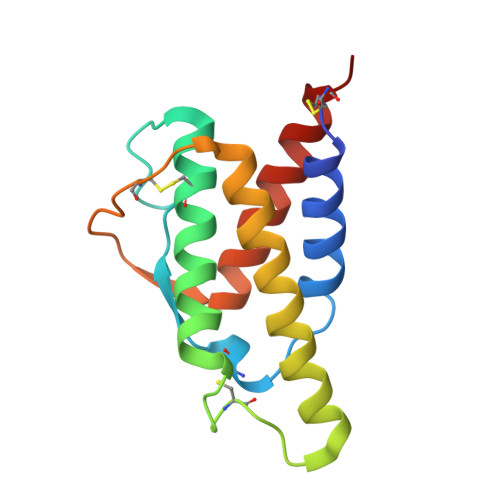
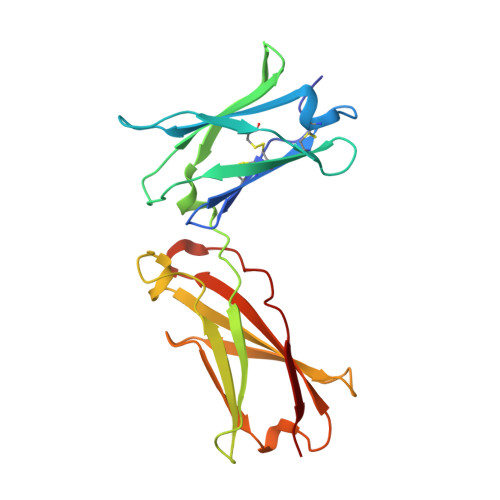
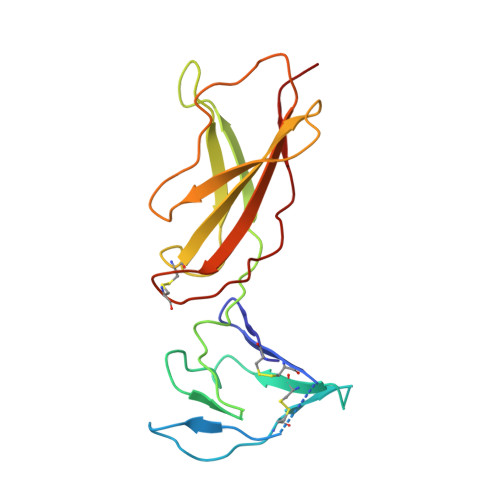
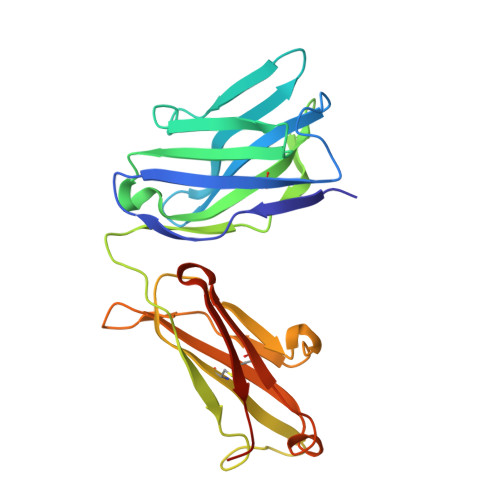
A strategy for the selection of monovalent antibodies that span protein dimer interfaces.
Spangler, J.B., Moraga, I., Jude, K.M., Savvides, C.S., Garcia, K.C.(2019) J Biological Chem 294: 13876-13886
- PubMed: 31387945
- DOI: https://doi.org/10.1074/jbc.RA119.009213
- Primary Citation of Related Structures:
6OEL - PubMed Abstract:
Ligand-induced dimerization is the predominant mechanism through which secreted proteins activate cell surface receptors to transmit essential biological signals. Cytokines are a large class of soluble proteins that dimerize transmembrane receptors into precise signaling topologies, but there is a need for alternative, engineerable ligand scaffolds that specifically recognize and stabilize these protein interactions. Recombinant antibodies can potentially serve as robust and versatile platforms for cytokine complex stabilization, and their specificity allows for tunable modulation of dimerization equilibrium. Here, we devised an evolutionary strategy to isolate monovalent antibody fragments that bridge together two different receptor subunits in a cytokine-receptor complex, precisely as the receptors are disposed in their natural signaling orientations. To do this, we screened a naive antibody library against a stabilized ligand-receptor ternary complex that acted as a "molecular cast" of the natural receptor dimer conformation. Our selections elicited "stapler" single-chain variable fragments (scFvs) of antibodies that specifically engage the interleukin-4 receptor heterodimer. The 3.1 Å resolution crystal structure of one such stapler revealed that, as intended, this scFv recognizes a composite epitope between the two receptors as they are positioned in the complex. Extending our approach, we evolved a stapler scFv that specifically binds to and stabilizes the interface between the interleukin-2 cytokine and one of its receptor subunits, leading to a 15-fold enhancement in interaction affinity. This demonstration that scFvs can be selected to recognize epitopes that span protein interfaces presents new opportunities to engineer structurally defined antibodies for a broad range of research and therapeutic applications.
- Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California 94305 jamie.spangler@jhu.edu.
Organizational Affiliation: