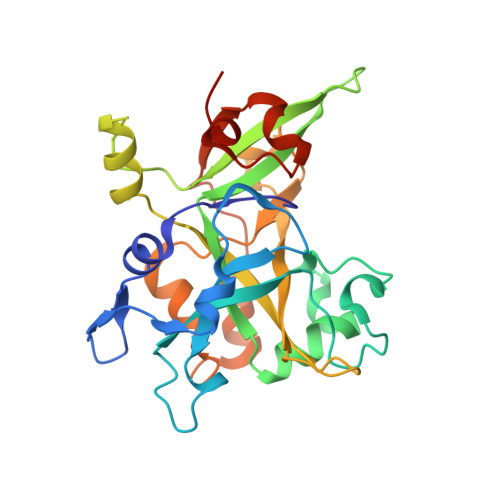
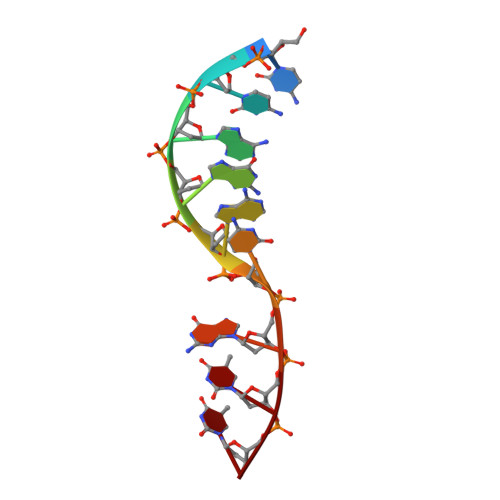
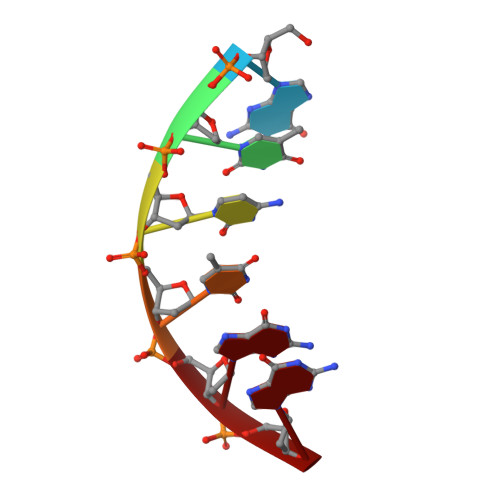
Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Halabelian, L., Ravichandran, M., Li, Y., Zeng, H., Rao, A., Aravind, L., Arrowsmith, C.H.(2019) Nat Struct Mol Biol 26: 607-612
- PubMed: 31235913
- DOI: https://doi.org/10.1038/s41594-019-0246-6
- Primary Citation of Related Structures:
5KO9, 6OE7, 6OEA, 6OEB - PubMed Abstract:
Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein (HMCES) can covalently cross-link to abasic sites in single-stranded DNA at stalled replication forks to prevent genome instability. Here, we report crystal structures of the human HMCES SOS response-associated peptidase (SRAP) domain in complex with DNA-damage substrates, including HMCES cross-linked with an abasic site within a 3' overhang DNA. HMCES interacts with both single-strand and duplex segments of DNA, with two independent duplex DNA interaction sites identified in the SRAP domain. The HMCES DNA-protein cross-link structure provides structural insights into a novel thiazolidine covalent interaction between the DNA abasic site and conserved Cys 2 of HMCES. Collectively, our structures demonstrate the capacity for the SRAP domain to interact with a variety of single-strand- and double-strand-containing DNA structures found in DNA-damage sites, including 5' and 3' overhang DNAs and gapped DNAs with short single-strand segments.
- Structural Genomics Consortium, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: