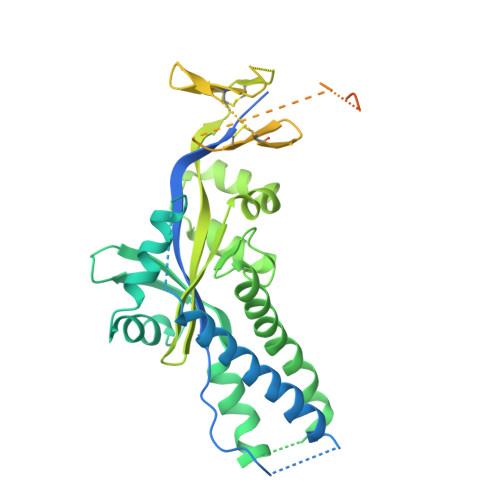
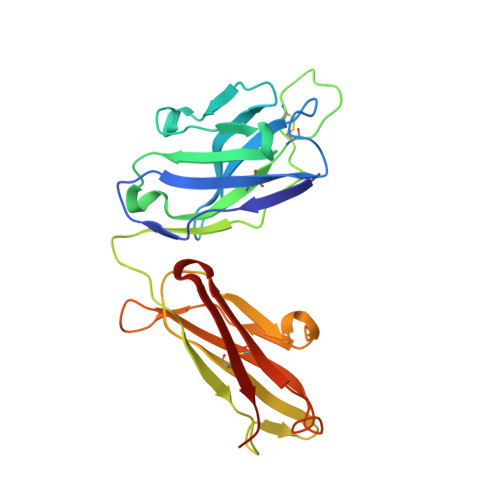
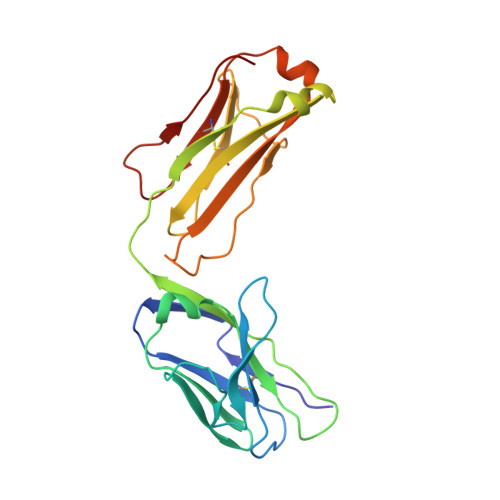
Transient opening of trimeric prefusion RSV F proteins.
Gilman, M.S.A., Furmanova-Hollenstein, P., Pascual, G., B van 't Wout, A., Langedijk, J.P.M., McLellan, J.S.(2019) Nat Commun 10: 2105-2105
- PubMed: 31068578
- DOI: https://doi.org/10.1038/s41467-019-09807-5
- Primary Citation of Related Structures:
6OE4, 6OE5 - PubMed Abstract:
The respiratory syncytial virus (RSV) F glycoprotein is a class I fusion protein that mediates viral entry and is a major target of neutralizing antibodies. Structures of prefusion forms of RSV F, as well as other class I fusion proteins, have revealed compact trimeric arrangements, yet whether these trimeric forms can transiently open remains unknown. Here, we perform structural and biochemical studies on a recently isolated antibody, CR9501, and demonstrate that it enhances the opening of prefusion-stabilized RSV F trimers. The 3.3 Å crystal structure of monomeric RSV F bound to CR9501, combined with analysis of over 25 previously determined RSV F structures, reveals a breathing motion of the prefusion conformation. We also demonstrate that full-length RSV F trimers transiently open and dissociate on the cell surface. Collectively, these findings have implications for the function of class I fusion proteins, as well as antibody prophylaxis and vaccine development for RSV.
- Department of Biochemistry and Cell Biology, Geisel School of Medicine at Dartmouth, Hanover, NH, 03755, USA.
Organizational Affiliation: