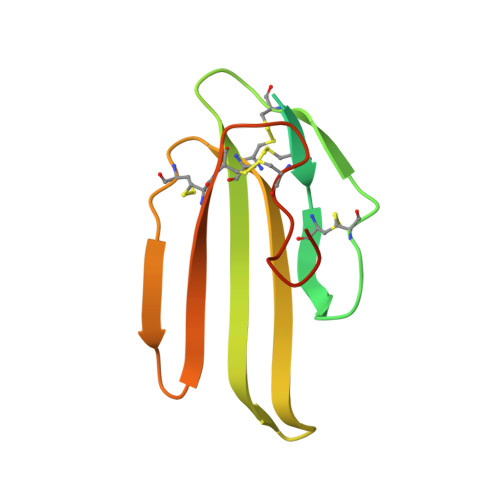
Structure of lipoprotein lipase in complex with GPIHBP1.
Arora, R., Nimonkar, A.V., Baird, D., Wang, C., Chiu, C.H., Horton, P.A., Hanrahan, S., Cubbon, R., Weldon, S., Tschantz, W.R., Mueller, S., Brunner, R., Lehr, P., Meier, P., Ottl, J., Voznesensky, A., Pandey, P., Smith, T.M., Stojanovic, A., Flyer, A., Benson, T.E., Romanowski, M.J., Trauger, J.W.(2019) Proc Natl Acad Sci U S A 116: 10360-10365
- PubMed: 31072929
- DOI: https://doi.org/10.1073/pnas.1820171116
- Primary Citation of Related Structures:
6OAU, 6OAZ, 6OB0 - PubMed Abstract:
Lipoprotein lipase (LPL) plays a central role in triglyceride (TG) metabolism. By catalyzing the hydrolysis of TGs present in TG-rich lipoproteins (TRLs), LPL facilitates TG utilization and regulates circulating TG and TRL concentrations. Until very recently, structural information for LPL was limited to homology models, presumably due to the propensity of LPL to unfold and aggregate. By coexpressing LPL with a soluble variant of its accessory protein glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1 (GPIHBP1) and with its chaperone protein lipase maturation factor 1 (LMF1), we obtained a stable and homogenous LPL/GPIHBP1 complex that was suitable for structure determination. We report here X-ray crystal structures of human LPL in complex with human GPIHBP1 at 2.5-3.0 Å resolution, including a structure with a novel inhibitor bound to LPL. Binding of the inhibitor resulted in ordering of the LPL lid and lipid-binding regions and thus enabled determination of the first crystal structure of LPL that includes these important regions of the protein. It was assumed for many years that LPL was only active as a homodimer. The structures and additional biochemical data reported here are consistent with a new report that LPL, in complex with GPIHBP1, can be active as a monomeric 1:1 complex. The crystal structures illuminate the structural basis for LPL-mediated TRL lipolysis as well as LPL stabilization and transport by GPIHBP1.
- Chemical Biology and Therapeutics, Novartis Institutes for Biomedical Research, Cambridge, MA 02139.
Organizational Affiliation: