An engineered antibody fragment targeting mutant beta-catenin via major histocompatibility complex I neoantigen presentation.
Miller, M.S., Douglass, J., Hwang, M.S., Skora, A.D., Murphy, M., Papadopoulos, N., Kinzler, K.W., Vogelstein, B., Zhou, S., Gabelli, S.B.(2019) J Biological Chem 294: 19322-19334
- PubMed: 31690625
- DOI: https://doi.org/10.1074/jbc.RA119.010251
- Primary Citation of Related Structures:
6O9B, 6O9C - PubMed Abstract:
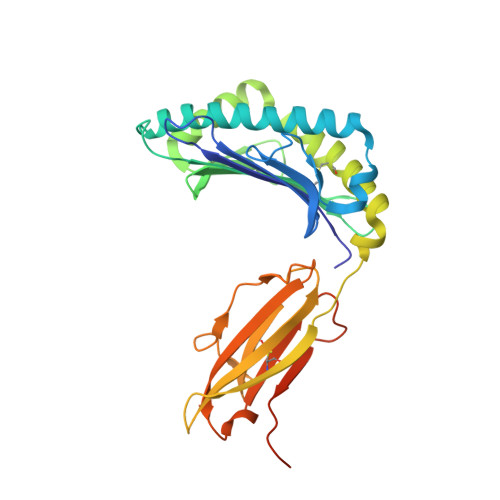
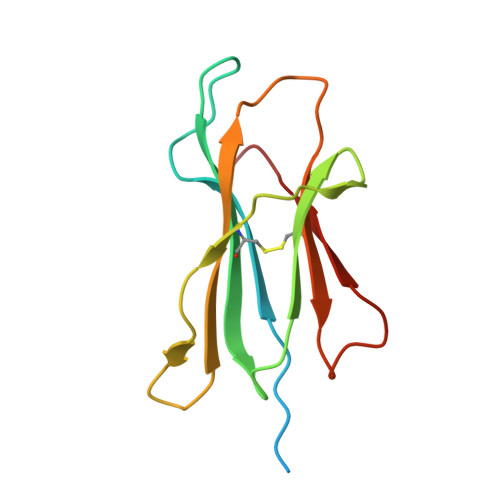
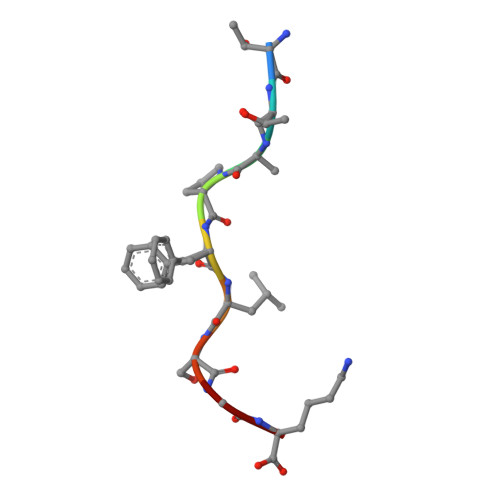
Mutations in CTNNB1 , the gene encoding β-catenin, are common in colon and liver cancers, the most frequent mutation affecting Ser-45 in β-catenin. Peptides derived from WT β-catenin have previously been shown to be presented on the cell surface as part of major histocompatibility complex (MHC) class I, suggesting an opportunity for targeting this common driver gene mutation with antibody-based therapies. Here, crystal structures of both the WT and S45F mutant peptide bound to HLA-A*03:01 at 2.20 and 2.45 Å resolutions, respectively, confirmed the accessibility of the phenylalanine residue for antibody recognition. Phage display was then used to identify single-chain variable fragment clones that selectively bind the S45F mutant peptide presented in HLA-A*03:01 and have minimal WT or other off-target binding. Following the initial characterization of five clones, we selected a single clone, E10, for further investigation. We developed a computational model of the binding of E10 to the mutant peptide-bound HLA-A3, incorporating data from affinity maturation as initial validation. In the future, our model may be used to design clones with maintained specificity and higher affinity. Such derivatives could be adapted into either cell-based (CAR-T) or protein-based (bispecific T-cell engagers) therapies to target cancer cells harboring the S45F mutation in CTNNB1 .
- Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland 21287.
Organizational Affiliation: