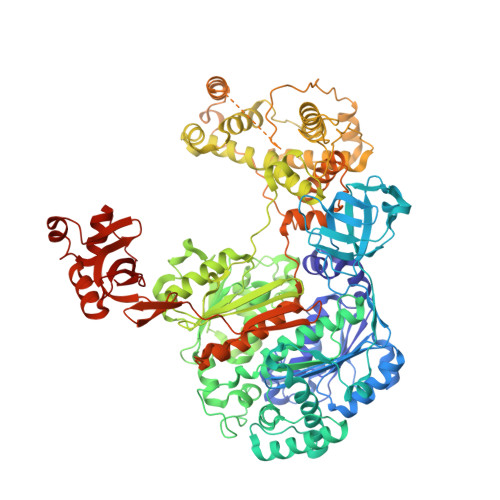
Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Hann, Z.S., Ji, C., Olsen, S.K., Lu, X., Lux, M.C., Tan, D.S., Lima, C.D.(2019) Proc Natl Acad Sci U S A 116: 15475-15484
- PubMed: 31235585
- DOI: https://doi.org/10.1073/pnas.1905488116
- Primary Citation of Related Structures:
6O82, 6O83 - PubMed Abstract:
The ubiquitin (Ub) and Ub-like (Ubl) protein-conjugation cascade is initiated by E1 enzymes that catalyze Ub/Ubl activation through C-terminal adenylation, thioester bond formation with an E1 catalytic cysteine, and thioester bond transfer to Ub/Ubl E2 conjugating enzymes. Each of these reactions is accompanied by conformational changes of the E1 domain that contains the catalytic cysteine (Cys domain). Open conformations of the Cys domain are associated with adenylation and thioester transfer to E2s, while a closed conformation is associated with pyrophosphate release and thioester bond formation. Several structures are available for Ub E1s, but none has been reported in the open state before pyrophosphate release or in the closed state. Here, we describe the structures of Schizosaccharomyces pombe Ub E1 in these two states, captured using semisynthetic Ub probes. In the first, with a Ub-adenylate mimetic (Ub-AMSN) bound, the E1 is in an open conformation before release of pyrophosphate. In the second, with a Ub-vinylsulfonamide (Ub-AVSN) bound covalently to the catalytic cysteine, the E1 is in a closed conformation required for thioester bond formation. These structures provide further insight into Ub E1 adenylation and thioester bond formation. Conformational changes that accompany Cys-domain rotation are conserved for SUMO and Ub E1s, but changes in Ub E1 involve additional surfaces as mutational and biochemical analysis of residues within these surfaces alter Ub E1 activities.
- Structural Biology Program, Sloan Kettering Institute, New York, NY 10065.
Organizational Affiliation: