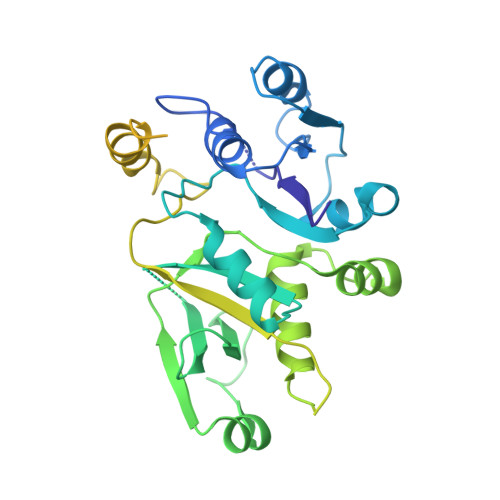
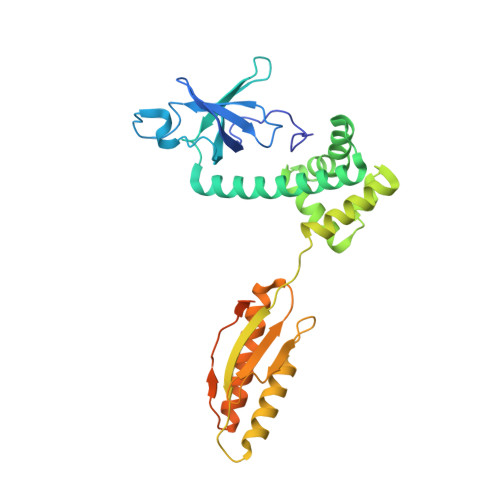
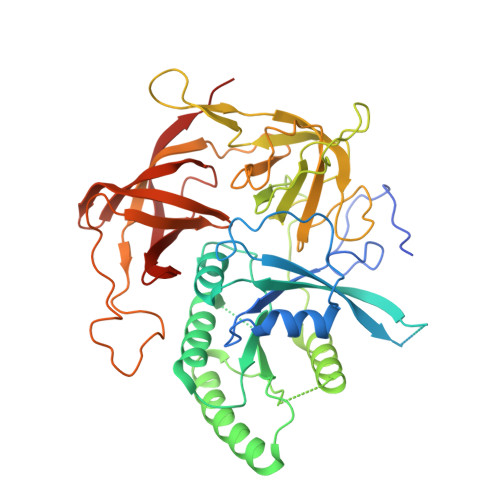
eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.
Kenner, L.R., Anand, A.A., Nguyen, H.C., Myasnikov, A.G., Klose, C.J., McGeever, L.A., Tsai, J.C., Miller-Vedam, L.E., Walter, P., Frost, A.(2019) Science 364: 491-495
- PubMed: 31048491
- DOI: https://doi.org/10.1126/science.aaw2922
- Primary Citation of Related Structures:
6O81, 6O85, 6O9Z - PubMed Abstract:
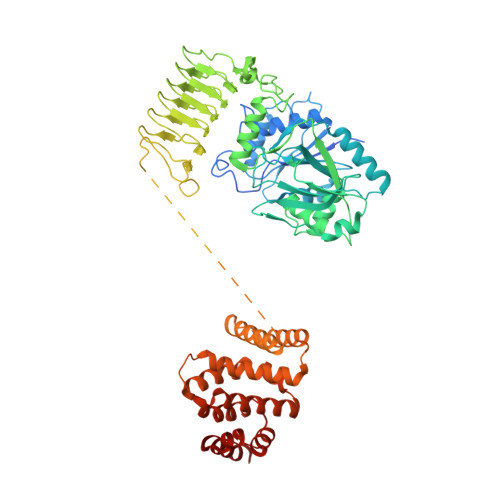
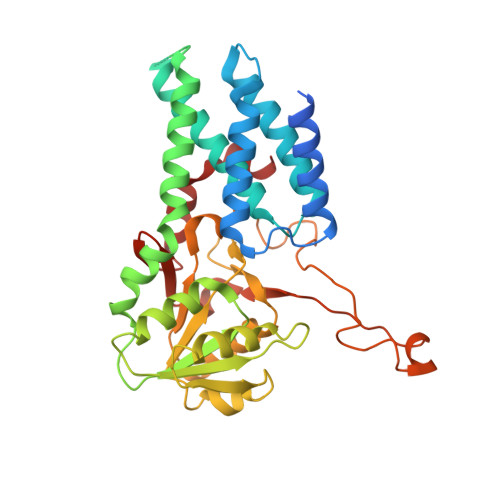
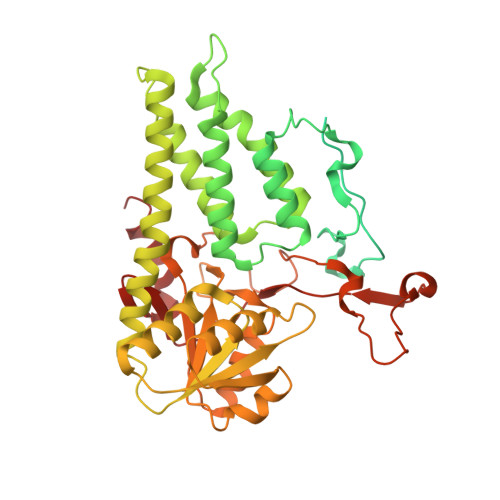
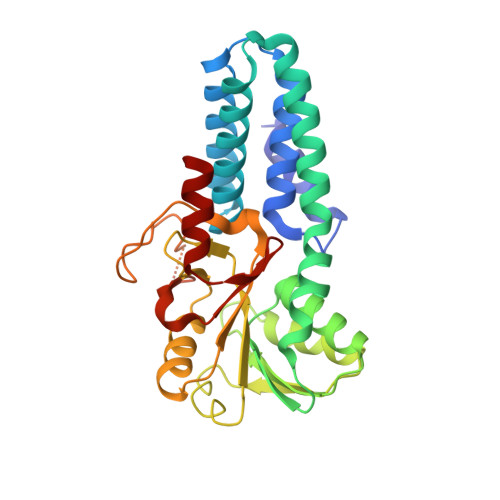
The integrated stress response (ISR) tunes the rate of protein synthesis. Control is exerted by phosphorylation of the general translation initiation factor eIF2. eIF2 is a guanosine triphosphatase that becomes activated by eIF2B, a two-fold symmetric and heterodecameric complex that functions as eIF2's dedicated nucleotide exchange factor. Phosphorylation converts eIF2 from a substrate into an inhibitor of eIF2B. We report cryo-electron microscopy structures of eIF2 bound to eIF2B in the dephosphorylated state. The structures reveal that the eIF2B decamer is a static platform upon which one or two flexible eIF2 trimers bind and align with eIF2B's bipartite catalytic centers to catalyze nucleotide exchange. Phosphorylation refolds eIF2α, allowing it to contact eIF2B at a different interface and, we surmise, thereby sequestering it into a nonproductive complex.
- Department of Biochemistry and Biophysics, University of California, San Francisco, CA, USA.
Organizational Affiliation: