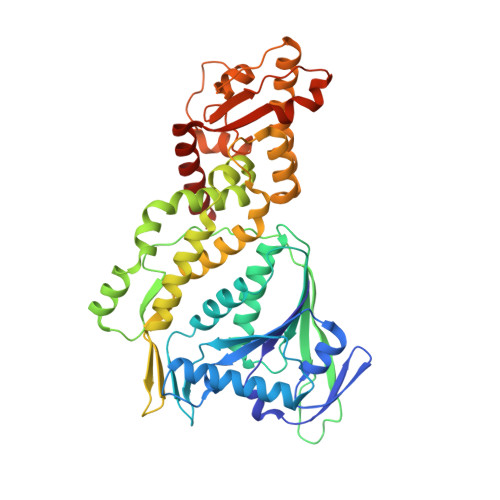
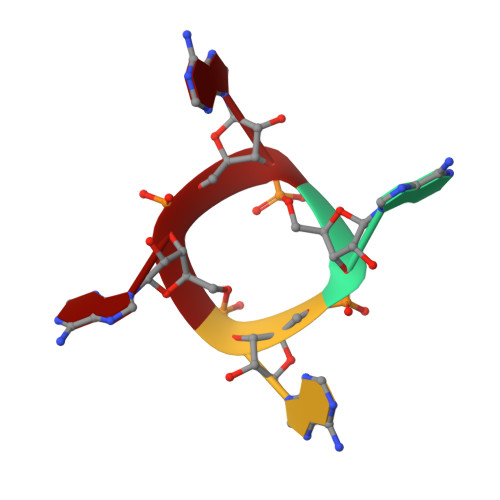
CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Jia, N., Jones, R., Yang, G., Ouerfelli, O., Patel, D.J.(2019) Mol Cell 75: 944-956.e6
- PubMed: 31326273
- DOI: https://doi.org/10.1016/j.molcel.2019.06.014
- Primary Citation of Related Structures:
6O6S, 6O6T, 6O6V, 6O6X, 6O6Y, 6O6Z, 6O70, 6O71, 6OV0 - PubMed Abstract:
Type III-A CRISPR-Cas surveillance complexes containing multi-subunit Csm effector, guide, and target RNAs exhibit multiple activities, including formation of cyclic-oligoadenylates (cA n ) from ATP and subsequent cA n -mediated cleavage of single-strand RNA (ssRNA) by the trans-acting Csm6 RNase. Our structure-function studies have focused on Thermococcus onnurineus Csm6 to deduce mechanistic insights into how cA 4 binding to the Csm6 CARF domain triggers the RNase activity of the Csm6 HEPN domain and what factors contribute to regulation of RNA cleavage activity. We demonstrate that the Csm6 CARF domain is a ring nuclease, whereby bound cA 4 is stepwise cleaved initially to ApApApA>p and subsequently to ApA>p in its CARF domain-binding pocket, with such cleavage bursts using a timer mechanism to regulate the RNase activity of the Csm6 HEPN domain. In addition, we establish T. onnurineus Csm6 as an adenosine-specific RNase and identify a histidine in the cA 4 CARF-binding pocket involved in autoinhibitory regulation of RNase activity.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA. Electronic address: jian@mskcc.org.
Organizational Affiliation: