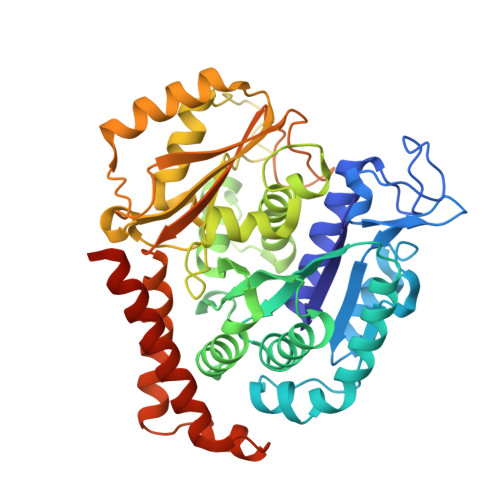
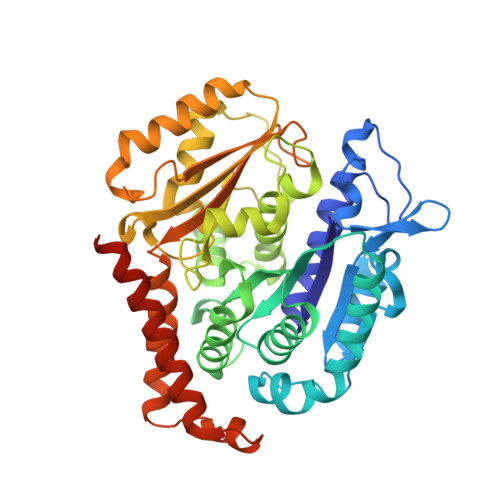
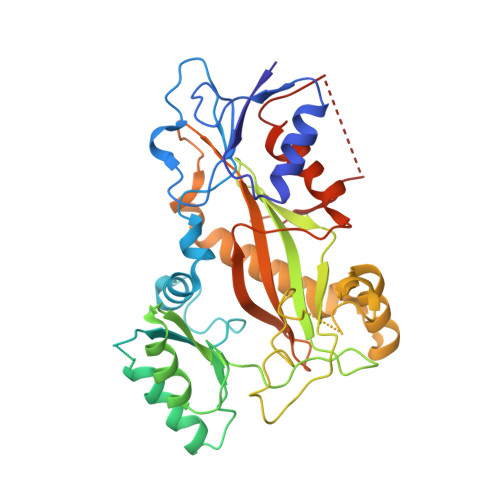
Structure-Guided Design, Synthesis, and Biological Evaluation of (2-(1H-Indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) Methanone (ABI-231) Analogues Targeting the Colchicine Binding Site in Tubulin.
Wang, Q., Arnst, K.E., Wang, Y., Kumar, G., Ma, D., White, S.W., Miller, D.D., Li, W., Li, W.(2019) J Med Chem 62: 6734-6750
- PubMed: 31251599
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00706
- Primary Citation of Related Structures:
6O5M, 6O5N, 6O61 - PubMed Abstract:
ABI-231 is a potent, orally bioavailable tubulin inhibitor that interacts with the colchicine binding site and is currently undergoing clinical trials for prostate cancer. Guided by the crystal structure of ABI-231 in complex with tubulin, we performed structure-activity relationship studies around the 3-indole moiety that led to the discovery of several potent ABI-231 analogues, most notably 10ab and 10bb . The crystal structures of 10ab and 10bb in complex with tubulin confirmed their improved molecular interactions to the colchicine site. In vitro, biological studies showed that new ABI-231 analogues disrupt tubulin polymerization, promote microtubule fragmentation, and inhibit cancer cell migration. In vivo, analogue 10bb not only significantly inhibits primary tumor growth and decreases tumor metastasis in melanoma xenograft models but also shows a significant ability to overcome paclitaxel resistance in a taxane-resistant PC-3/TxR model. In addition, pharmacological screening suggested that 10bb has a low risk of potential off-target function.
- Department of Pharmaceutical Sciences, College of Pharmacy , University of Tennessee Health Science Center , Memphis , Tennessee 38163 , United States.
Organizational Affiliation: