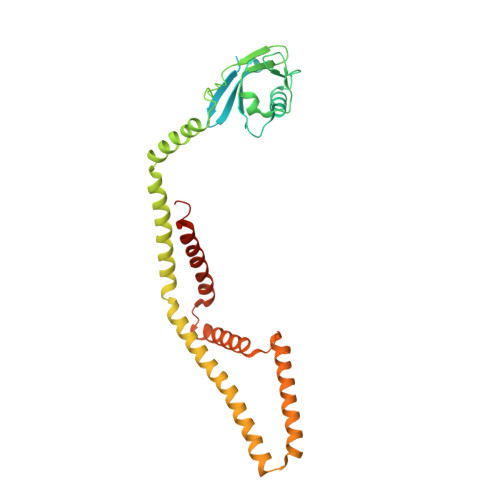
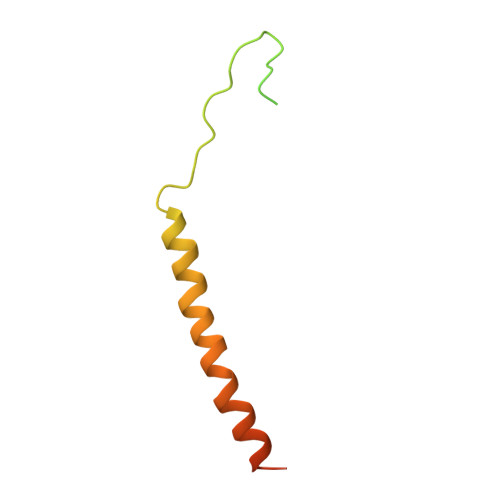
Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Wang, Y., Nguyen, N.X., She, J., Zeng, W., Yang, Y., Bai, X.C., Jiang, Y.(2019) Cell 177: 1252
- PubMed: 31080062
- DOI: https://doi.org/10.1016/j.cell.2019.03.050
- Primary Citation of Related Structures:
6O58, 6O5B - PubMed Abstract:
Mitochondrial calcium uptake is crucial to the regulation of eukaryotic Ca 2+ homeostasis and is mediated by the mitochondrial calcium uniporter (MCU). While MCU alone can transport Ca 2+ in primitive eukaryotes, metazoans require an essential single membrane-spanning auxiliary component called EMRE to form functional channels; however, the molecular mechanism of EMRE regulation remains elusive. Here, we present the cryo-EM structure of the human MCU-EMRE complex, which defines the interactions between MCU and EMRE as well as pinpoints the juxtamembrane loop of MCU and extended linker of EMRE as the crucial elements in the EMRE-dependent gating mechanism among metazoan MCUs. The structure also features the dimerization of two MCU-EMRE complexes along an interface at the N-terminal domain (NTD) of human MCU that is a hotspot for post-translational modifications. Thus, the human MCU-EMRE complex, which constitutes the minimal channel components among metazoans, provides a framework for future mechanistic studies on MCU.
- Howard Hughes Medical Institute and Department of Physiology, University of Texas Southwestern Medical Center, Dallas, TX, USA; Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: