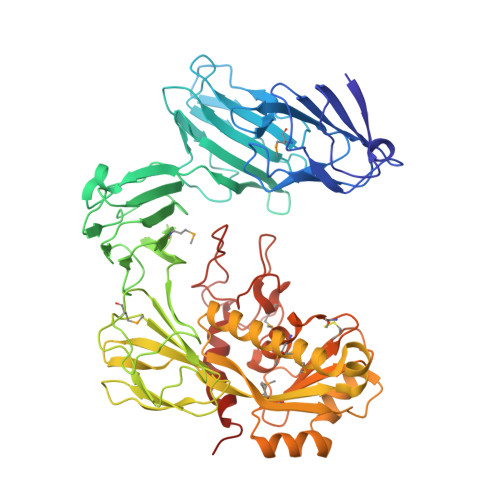
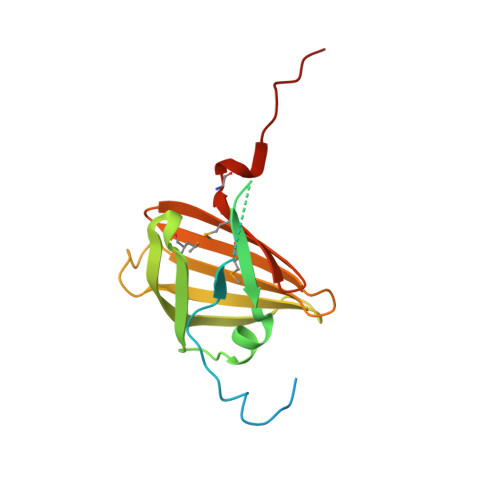
The structure ofAcinetobacter-secreted protease CpaA complexed with its chaperone CpaB reveals a novel mode of a T2SS chaperone-substrate interaction.
Urusova, D.V., Kinsella, R.L., Salinas, N.D., Haurat, M.F., Feldman, M.F., Tolia, N.H.(2019) J Biological Chem 294: 13344-13354
- PubMed: 31320476
- DOI: https://doi.org/10.1074/jbc.RA119.009805
- Primary Citation of Related Structures:
6O38 - PubMed Abstract:
Members of the Acinetobacter baumannii-calcoaceticus complex are nosocomial pathogens frequently causing multidrug-resistant infections that are increasing at alarming rates. A. baumannii has become the Gram-negative bacterium with the highest rate of multidrug resistance. As such, it is categorized by the World Health Organization as a critical priority for the research and development of new antimicrobial therapies. The zinc-dependent metalloendopeptidase CpaA is a predominant substrate of the type II secretion system (T2SS). CpaA is also a virulence factor of medically relevant Acinetobacter strains that specifically degrade the human glycoprotein coagulation factor XII and not its deglycosylated form, but the mechanism for this specificity is unknown. CpaB is a membrane-anchored T2SS chaperone that interacts with CpaA and is required for its stability and secretion. Here, we report the crystal structure of the CpaAB complex at 2.6-Å resolution, revealing four glycan-binding domains in CpaA that were not predicted from its primary sequence and may explain CpaA's glycoprotein-targeting activity. The structure of the complex identified a novel mode for chaperone-protease interactions in which the protease surrounds the chaperone. The CpaAB organization was akin to zymogen inactivation, with CpaB serving as a prodomain that inhibits catalytically active CpaA. CpaB contains a C-terminal tail that appears to block access to the CpaA catalytic site, and functional experiments with truncated variants indicated that this tail is dispensable for CpaA expression and secretion. Our results provide new insight into the mechanism of CpaA secretion and may inform the future development of therapeutic strategies for managing Acinetobacter infections.
- Department of Molecular Microbiology, Washington University School of Medicine, Campus Box 8230, Saint Louis, Missouri 63110.
Organizational Affiliation: