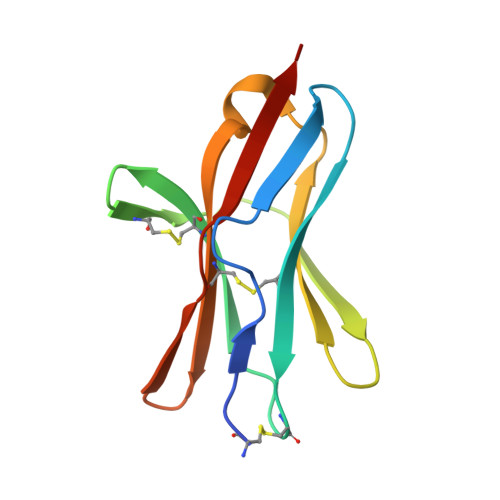
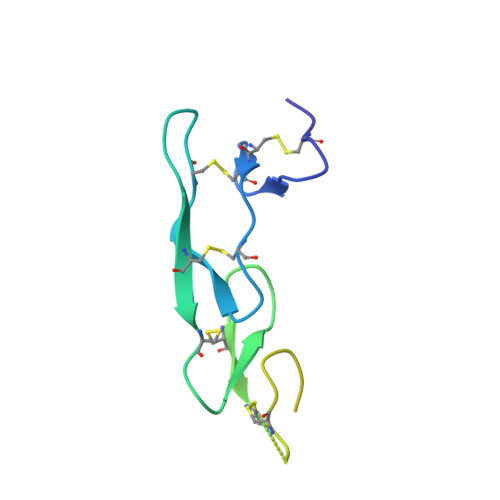
Structure of human cytomegalovirus UL144, an HVEM orthologue, bound to the B and T cell lymphocyte attenuator.
Bitra, A., Nemcovicova, I., Picarda, G., Doukov, T., Wang, J., Benedict, C.A., Zajonc, D.M.(2019) J Biological Chem 294: 10519-10529
- PubMed: 31126984
- DOI: https://doi.org/10.1074/jbc.RA119.009199
- Primary Citation of Related Structures:
6NYP - PubMed Abstract:
Human cytomegalovirus (HCMV) is a β-herpesvirus that has co-evolved with the host immune system to establish lifelong persistence. HCMV encodes many immunomodulatory molecules, including the glycoprotein UL144. UL144 is a structural mimic of the tumor necrosis factor receptor superfamily member HVEM (herpesvirus entry mediator), which binds to the various ligands LIGHT, LTα, BTLA, CD160, and gD. However, in contrast to HVEM, UL144 only binds BTLA, inhibiting T-cell activation. Here, we report the crystal structure of the UL144-BTLA complex, revealing that UL144 utilizes residues from its N-terminal cysteine-rich domain 1 (CRD1) to interact uniquely with BTLA. The shorter CRD2 loop of UL144 also alters the relative orientation of BTLA binding with both N-terminal CRDs. By employing structure-guided mutagenesis, we have identified a mutant of BTLA (L123A) that interferes with HVEM binding but preserves UL144 interactions. Furthermore, our results illuminate structural differences between UL144 and HVEM that explain its binding selectivity and highlight it as a suitable scaffold for designing superior, immune inhibitory BTLA agonists.
- From the Division of Immune Regulation, La Jolla Institute for Immunology, La Jolla, California 92037.
Organizational Affiliation: