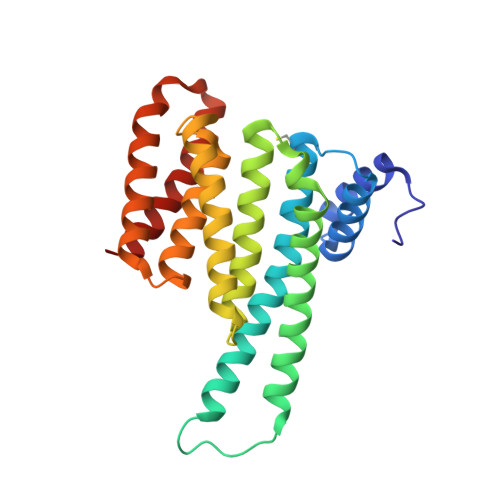
Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
Wolter, M., de Vink, P., Neves, J.F., Srdanovic, S., Higuchi, Y., Kato, N., Wilson, A., Landrieu, I., Brunsveld, L., Ottmann, C.(2020) J Am Chem Soc 142: 11772-11783
- PubMed: 32501683
- DOI: https://doi.org/10.1021/jacs.0c02151
- Primary Citation of Related Structures:
6NV2, 6QHL, 6QHM - PubMed Abstract:
Natural compounds are an important class of potent drug molecules including some retrospectively found to act as stabilizers of protein-protein interactions (PPIs). However, the design of synthetic PPI stabilizers remains an understudied approach. To date, there are limited examples where cooperativity has been utilized to guide the optimization of a PPI stabilizer. The 14-3-3 scaffold proteins provide an excellent platform to explore PPI stabilization because these proteins mediate several hundred PPIs, and a class of natural compounds, the fusicoccanes, are known to stabilize a subset of 14-3-3 protein interactions. 14-3-3 has been reported to negatively regulate the p65 subunit of the NF-κB transcription factor, which qualifies this protein complex as a potential target for drug discovery to control cell proliferation. Here, we report the high-resolution crystal structures of two 14-3-3 binding motifs of p65 in complex with 14-3-3. A semisynthetic natural product derivative, DP-005, binds to an interface pocket of the p65/14-3-3 complex and concomitantly stabilizes it. Cooperativity analyses of this interaction, and other disease relevant 14-3-3-PPIs, demonstrated selectivity of DP-005 for the p65/14-3-3 complex. The adaptation of a cooperative binding model provided a general approach to characterize stabilization and to assay for selectivity of PPI stabilizers.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Technische Universiteit Eindhoven, P.O. Box 513, Eindhoven 5600 MB, The Netherlands.
Organizational Affiliation: