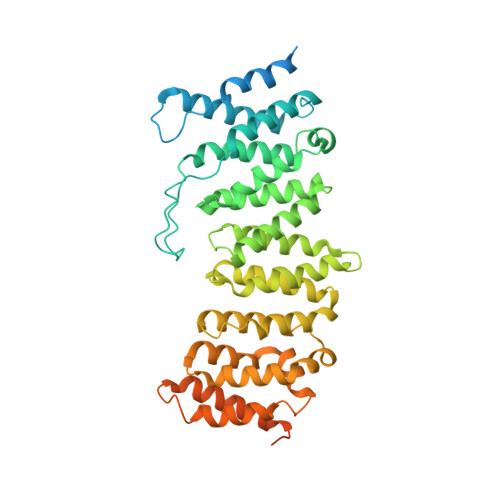
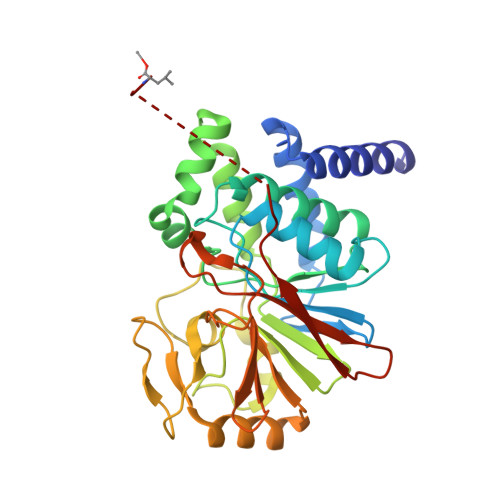
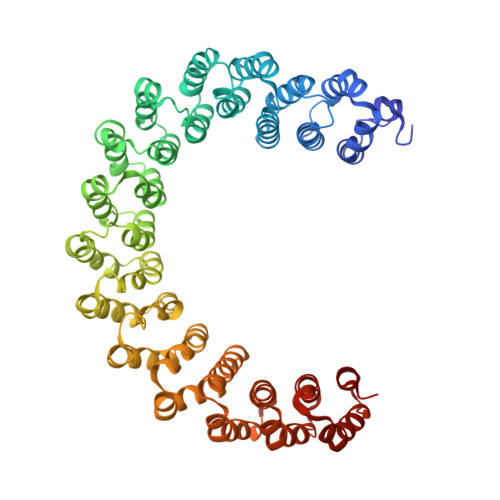Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Leonard, D., Huang, W., Izadmehr, S., O'Connor, C.M., Wiredja, D.D., Wang, Z., Zaware, N., Chen, Y., Schlatzer, D.M., Kiselar, J., Vasireddi, N., Schuchner, S., Perl, A.L., Galsky, M.D., Xu, W., Brautigan, D.L., Ogris, E., Taylor, D.J., Narla, G.(2020) Cell 181: 688-701.e16
- PubMed: 32315618
- DOI: https://doi.org/10.1016/j.cell.2020.03.038
- Primary Citation of Related Structures:
6NTS - PubMed Abstract:
Impairment of protein phosphatases, including the family of serine/threonine phosphatases designated PP2A, is essential for the pathogenesis of many diseases, including cancer. The ability of PP2A to dephosphorylate hundreds of proteins is regulated by over 40 specificity-determining regulatory "B" subunits that compete for assembly and activation of heterogeneous PP2A heterotrimers. Here, we reveal how a small molecule, DT-061, specifically stabilizes the B56α-PP2A holoenzyme in a fully assembled, active state to dephosphorylate selective substrates, such as its well-known oncogenic target, c-Myc. Our 3.6 Å structure identifies molecular interactions between DT-061 and all three PP2A subunits that prevent dissociation of the active enzyme and highlight inherent mechanisms of PP2A complex assembly. Thus, our findings provide fundamental insights into PP2A complex assembly and regulation, identify a unique interfacial stabilizing mode of action for therapeutic targeting, and aid in the development of phosphatase-based therapeutics tailored against disease specific phospho-protein targets.
- Department of Pathology, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA; Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH 44106, USA.
Organizational Affiliation: