BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
Herrou, J., Willett, J.W., Fiebig, A., Czyz, D.M., Cheng, J.X., Ultee, E., Briegel, A., Bigelow, L., Babnigg, G., Kim, Y., Crosson, S.(2019) J Bacteriol 201
- PubMed: 30936371
- DOI: https://doi.org/10.1128/JB.00134-19
- Primary Citation of Related Structures:
6NTR - PubMed Abstract:
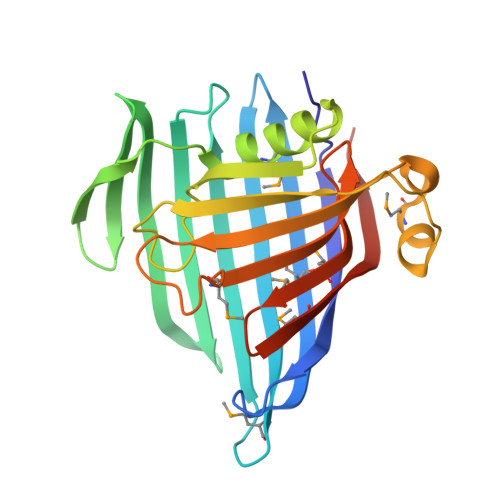
The Gram-negative cell envelope is a remarkable structure with core components that include an inner membrane, an outer membrane, and a peptidoglycan layer in the periplasmic space between. Multiple molecular systems function to maintain integrity of this essential barrier between the interior of the cell and its surrounding environment. We show that a conserved DUF1849 family protein, EipB, is secreted to the periplasmic space of Brucella species, a monophyletic group of intracellular pathogens. In the periplasm, EipB folds into an unusual 14-stranded β-spiral structure that resembles the LolA and LolB lipoprotein delivery system, though the overall fold of EipB is distinct from LolA/LolB. Deletion of eipB results in defects in Brucella cell envelope integrity in vitro and in maintenance of spleen colonization in a mouse model of Brucella abortus infection. Transposon disruption of ttpA , which encodes a periplasmic protein containing tetratricopeptide repeats, is synthetically lethal with eipB deletion. ttpA is a reported virulence determinant in Brucella , and our studies of ttpA deletion and overexpression strains provide evidence that this gene also contributes to cell envelope function. We conclude that eipB and ttpA function in the Brucella periplasmic space to maintain cell envelope integrity, which facilitates survival in a mammalian host. IMPORTANCE Brucella species cause brucellosis, a global zoonosis. A gene encoding a conserved DUF1849-family protein, which we have named EipB, is present in all sequenced Brucella and several other genera in the class Alphaproteobacteria The manuscript provides the first functional and structural characterization of a DUF1849 protein. We show that EipB is secreted to the periplasm where it forms a spiral-shaped antiparallel β protein that is a determinant of cell envelope integrity in vitro and virulence in an animal model of disease. eipB genetically interacts with ttpA , which also encodes a periplasmic protein. We propose that EipB and TtpA function as part of a system required for cell envelope homeostasis in select Alphaproteobacteria .
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, Illinois, USA.
Organizational Affiliation: