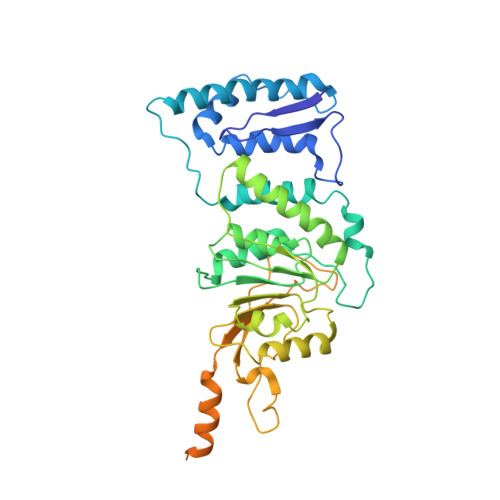
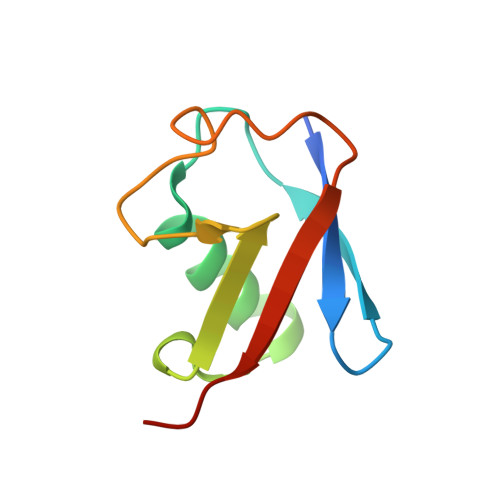
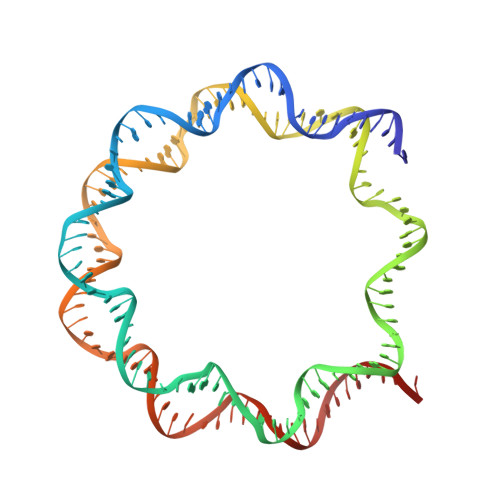
Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase.
Anderson, C.J., Baird, M.R., Hsu, A., Barbour, E.H., Koyama, Y., Borgnia, M.J., McGinty, R.K.(2019) Cell Rep 26: 1681-1690.e5
- PubMed: 30759380
- DOI: https://doi.org/10.1016/j.celrep.2019.01.058
- Primary Citation of Related Structures:
6NN6 - PubMed Abstract:
Histone H3 lysine 79 (H3K79) methylation is enriched on actively transcribed genes, and its misregulation is a hallmark of leukemia. Methylation of H3K79, which resides on the structured disk face of the nucleosome, is mediated by the Dot1L methyltransferase. Dot1L activity is part of a trans-histone crosstalk pathway, requiring prior histone H2B ubiquitylation of lysine 120 (H2BK120ub) for optimal activity. However, the molecular details describing both how Dot1L binds to the nucleosome and why Dot1L is activated by H2BK120 ubiquitylation are unknown. Here, we present the cryoelectron microscopy (cryo-EM) structure of Dot1L bound to a nucleosome reconstituted with site-specifically ubiquitylated H2BK120. The structure reveals that Dot1L engages the nucleosome acidic patch using a variant arginine anchor and occupies a conformation poised for methylation. In this conformation, Dot1L and ubiquitin interact directly through complementary hydrophobic surfaces. This study establishes a path to better understand Dot1L function in normal and leukemia cells.
- Department of Biochemistry and Biophysics, School of Medicine, The University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Organizational Affiliation: