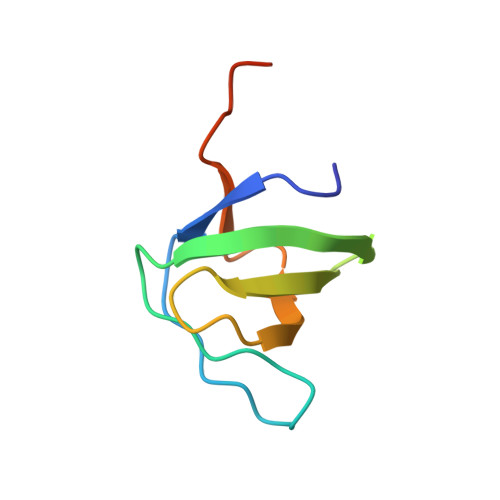
Crystal structure of the SH3 domain of human Lyn non-receptor tyrosine kinase.
Berndt, S., Gurevich, V.V., Iverson, T.M.(2019) PLoS One 14: e0215140-e0215140
- PubMed: 30969999
- DOI: https://doi.org/10.1371/journal.pone.0215140
- Primary Citation of Related Structures:
6NMW - PubMed Abstract:
Lyn kinase (Lck/Yes related novel protein tyrosine kinase) belongs to the family of Src-related non-receptor tyrosine kinases. Consistent with physiological roles in cell growth and proliferation, aberrant function of Lyn is associated with various forms of cancer, including leukemia, breast cancer and melanoma. Here, we determine a 1.3 Å resolution crystal structure of the polyproline-binding SH3 regulatory domain of human Lyn kinase, which adopts a five-stranded β-barrel fold. Mapping of cancer-associated point mutations onto this structure reveals that these amino acid substitutions are distributed throughout the SH3 domain and may affect Lyn kinase function distinctly.
- Department of Pharmacology, Vanderbilt University, Nashville, TN, United States of America.
Organizational Affiliation: