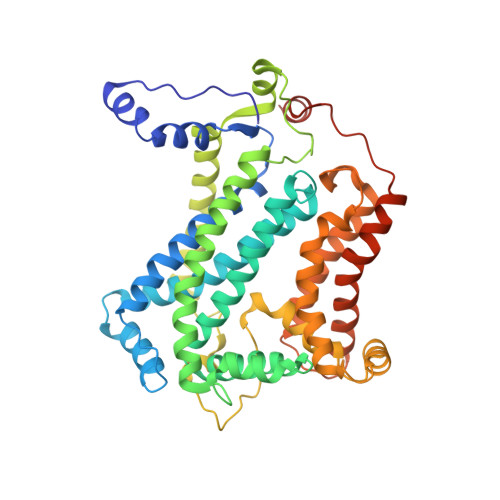
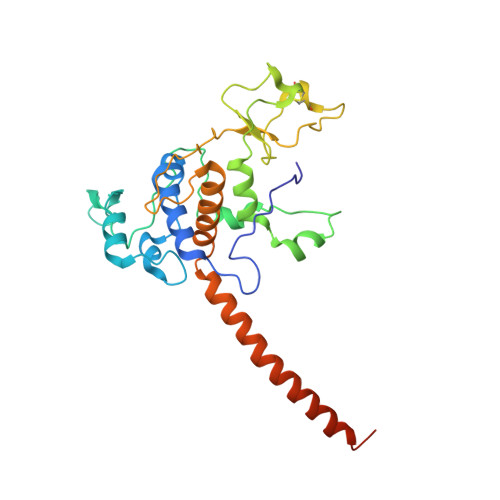
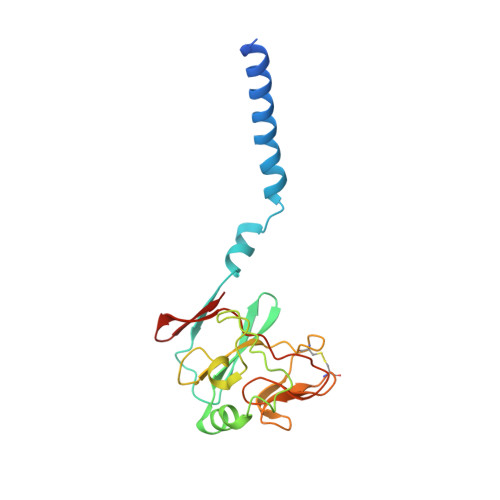
Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit.
Esser, L., Zhou, F., Yu, C.A., Xia, D.(2019) J Biological Chem 294: 12007-12019
- PubMed: 31182483
- DOI: https://doi.org/10.1074/jbc.RA119.008381
- Primary Citation of Related Structures:
6NHG, 6NHH, 6NIN - PubMed Abstract:
Cytochrome bc 1 complexes (cyt bc 1 ), also known as complex III in mitochondria, are components of the cellular respiratory chain and of the photosynthetic apparatus of non-oxygenic photosynthetic bacteria. They catalyze electron transfer (ET) from ubiquinol to cytochrome c and concomitantly translocate protons across the membrane, contributing to the cross-membrane potential essential for a myriad of cellular activities. This ET-coupled proton translocation reaction requires a gating mechanism that ensures bifurcated electron flow. Here, we report the observation of the Rieske iron-sulfur protein (ISP) in a mobile state, as revealed by the crystal structure of cyt bc 1 from the photosynthetic bacterium Rhodobacter sphaeroides in complex with the fungicide azoxystrobin. Unlike cyt bc 1 inhibitors stigmatellin and famoxadone that immobilize the ISP, azoxystrobin causes the ISP-ED to separate from the cyt b subunit and to remain in a mobile state. Analysis of anomalous scattering signals from the iron-sulfur cluster of the ISP suggests the existence of a trajectory for electron delivery. This work supports and solidifies the hypothesis that the bimodal conformation switch of the ISP provides a gating mechanism for bifurcated ET, which is essential to the Q-cycle mechanism of cyt bc 1 function.
- Laboratory of Cell Biology, Center for Cancer Research, NCI, National Institutes of Health, Bethesda, Maryland 20892.
Organizational Affiliation: