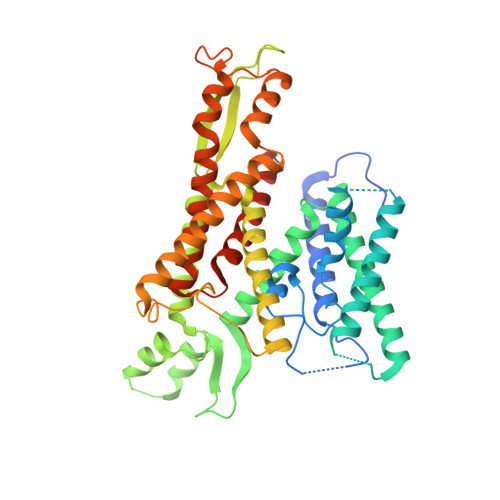
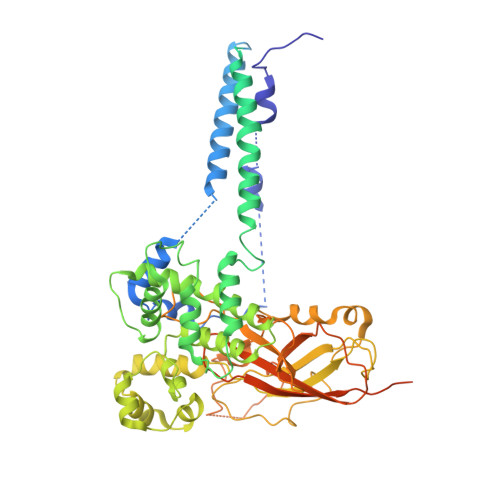
Structure of the post-translational protein translocation machinery of the ER membrane.
Wu, X., Cabanos, C., Rapoport, T.A.(2019) Nature 566: 136-139
- PubMed: 30644436
- DOI: https://doi.org/10.1038/s41586-018-0856-x
- Primary Citation of Related Structures:
6ND1 - PubMed Abstract:
Many proteins must translocate through the protein-conducting Sec61 channel in the eukaryotic endoplasmic reticulum membrane or the SecY channel in the prokaryotic plasma membrane 1,2 . Proteins with highly hydrophobic signal sequences are first recognized by the signal recognition particle (SRP) 3,4 and then moved co-translationally through the Sec61 or SecY channel by the associated translating ribosome. Substrates with less hydrophobic signal sequences bypass the SRP and are moved through the channel post-translationally 5,6 . In eukaryotic cells, post-translational translocation is mediated by the association of the Sec61 channel with another membrane protein complex, the Sec62-Sec63 complex 7-9 , and substrates are moved through the channel by the luminal BiP ATPase 9 . How the Sec62-Sec63 complex activates the Sec61 channel for post-translational translocation is not known. Here we report the electron cryo-microscopy structure of the Sec complex from Saccharomyces cerevisiae, consisting of the Sec61 channel and the Sec62, Sec63, Sec71 and Sec72 proteins. Sec63 causes wide opening of the lateral gate of the Sec61 channel, priming it for the passage of low-hydrophobicity signal sequences into the lipid phase, without displacing the channel's plug domain. Lateral channel opening is triggered by Sec63 interacting both with cytosolic loops in the C-terminal half of Sec61 and transmembrane segments in the N-terminal half of the Sec61 channel. The cytosolic Brl domain of Sec63 blocks ribosome binding to the channel and recruits Sec71 and Sec72, positioning them for the capture of polypeptides associated with cytosolic Hsp70 10 . Our structure shows how the Sec61 channel is activated for post-translational protein translocation.
- Howard Hughes Medical Institute, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: