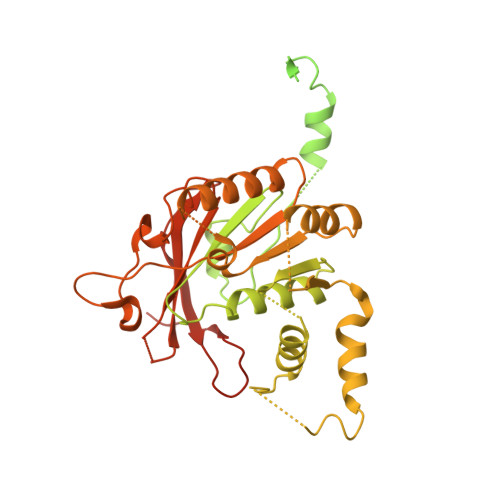
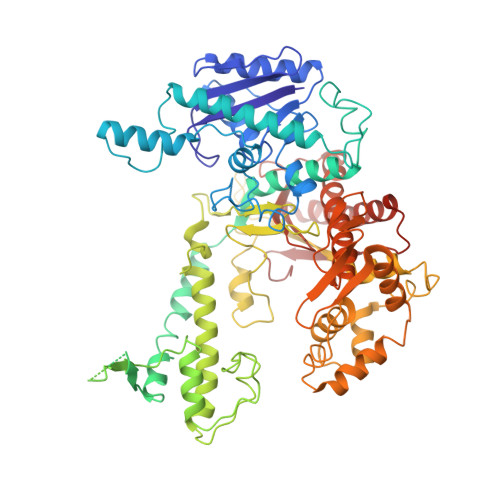
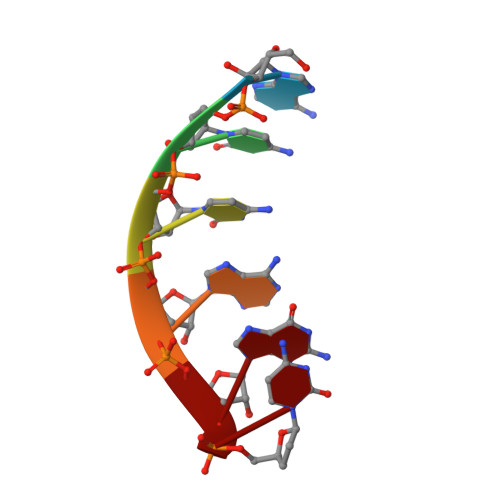
Structures and operating principles of the replisome.
Gao, Y., Cui, Y., Fox, T., Lin, S., Wang, H., de Val, N., Zhou, Z.H., Yang, W.(2019) Science 363
- PubMed: 30679383
- DOI: https://doi.org/10.1126/science.aav7003
- Primary Citation of Related Structures:
6N7I, 6N7N, 6N7S, 6N7T, 6N7V, 6N7W, 6N9U, 6N9V, 6N9W, 6N9X - PubMed Abstract:
Visualization in atomic detail of the replisome that performs concerted leading- and lagging-DNA strand synthesis at a replication fork has not been reported. Using bacteriophage T7 as a model system, we determined cryo-electron microscopy structures up to 3.2-angstroms resolution of helicase translocating along DNA and of helicase-polymerase-primase complexes engaging in synthesis of both DNA strands. Each domain of the spiral-shaped hexameric helicase translocates sequentially hand-over-hand along a single-stranded DNA coil, akin to the way AAA+ ATPases (adenosine triphosphatases) unfold peptides. Two lagging-strand polymerases are attached to the primase, ready for Okazaki fragment synthesis in tandem. A β hairpin from the leading-strand polymerase separates two parental DNA strands into a T-shaped fork, thus enabling the closely coupled helicase to advance perpendicular to the downstream DNA duplex. These structures reveal the molecular organization and operating principles of a replisome.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: