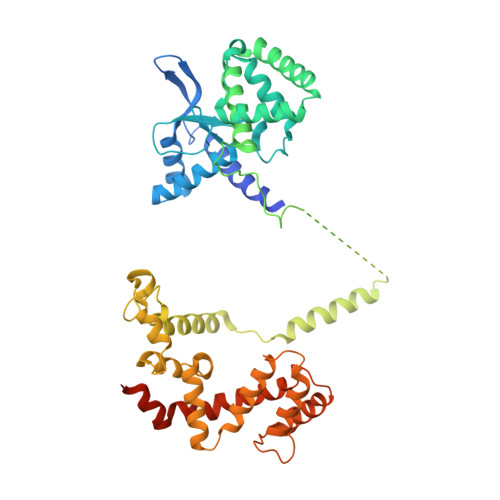
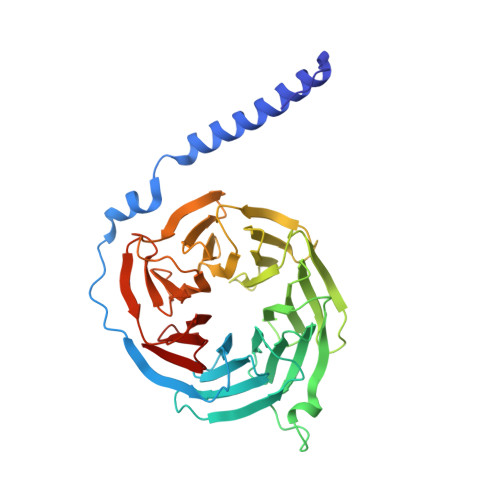
Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Patil, D.N., Rangarajan, E.S., Novick, S.J., Pascal, B.D., Kojetin, D.J., Griffin, P.R., Izard, T., Martemyanov, K.A.(2018) Elife 7
- PubMed: 30540250
- DOI: https://doi.org/10.7554/eLife.42150
- Primary Citation of Related Structures:
6N9G - PubMed Abstract:
Signaling by the G-protein-coupled receptors (GPCRs) plays fundamental role in a vast number of essential physiological functions. Precise control of GPCR signaling requires action of regulators of G protein signaling (RGS) proteins that deactivate heterotrimeric G proteins. RGS proteins are elaborately regulated and comprise multiple domains and subunits, yet structural organization of these assemblies is poorly understood. Here, we report a crystal structure and dynamics analyses of the multisubunit complex of RGS7, a major regulator of neuronal signaling with key roles in controlling a number of drug target GPCRs and links to neuropsychiatric disease, metabolism, and cancer. The crystal structure in combination with molecular dynamics and mass spectrometry analyses reveals unique organizational features of the complex and long-range conformational changes imposed by its constituent subunits during allosteric modulation. Notably, several intermolecular interfaces in the complex work in synergy to provide coordinated modulation of this key GPCR regulator.
- Department of Neuroscience, The Scripps Research Institute, Jupiter, United States.
Organizational Affiliation: