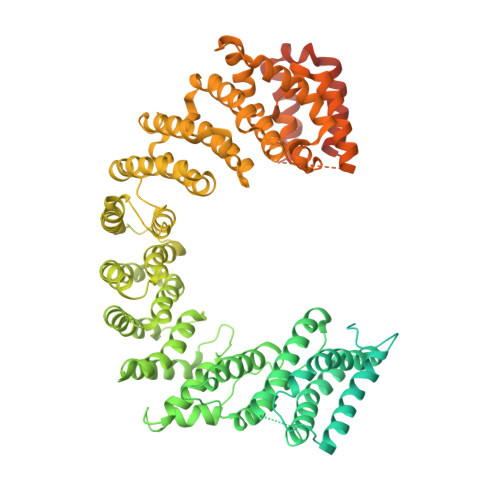
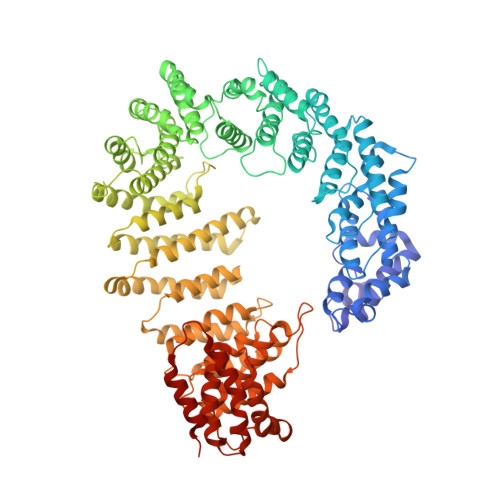
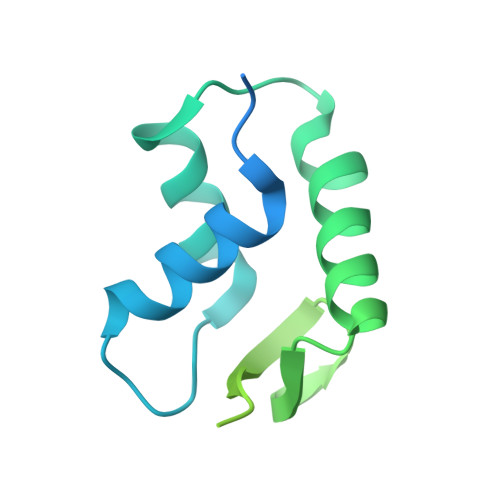
Fuzzy Interactions Form and Shape the Histone Transport Complex.
Ivic, N., Potocnjak, M., Solis-Mezarino, V., Herzog, F., Bilokapic, S., Halic, M.(2019) Mol Cell 73: 1191
- PubMed: 30824373
- DOI: https://doi.org/10.1016/j.molcel.2019.01.032
- Primary Citation of Related Structures:
6N88, 6N89 - PubMed Abstract:
Protein transport into the nucleus is mediated by transport receptors. Import of highly charged proteins, such as histone H1 and ribosomal proteins, requires a dimer of two transport receptors. In this study, we determined the cryo-EM structure of the Imp7:Impβ:H1.0 complex, showing that the two importins form a cradle that accommodates the linker histone. The H1.0 globular domain is bound to Impβ, whereas the acidic loops of Impβ and Imp7 chaperone the positively charged C-terminal tail. Although it remains disordered, the H1 tail serves as a zipper that closes and stabilizes the structure through transient non-specific interactions with importins. Moreover, we found that the GGxxF and FxFG motifs in the Imp7 C-terminal tail are essential for Imp7:Impβ dimerization and H1 import, resembling importin interaction with nucleoporins, which, in turn, promote complex disassembly. The architecture of many other complexes might be similarly defined by rapidly exchanging electrostatic interactions mediated by disordered regions.
- Gene Center Munich and Department of Biochemistry, Ludwig Maximilian University of Munich, 81377 Munich, Germany; Department of Physical Chemistry, Rudjer Boskovic Institute, 10000 Zagreb, Croatia.
Organizational Affiliation: