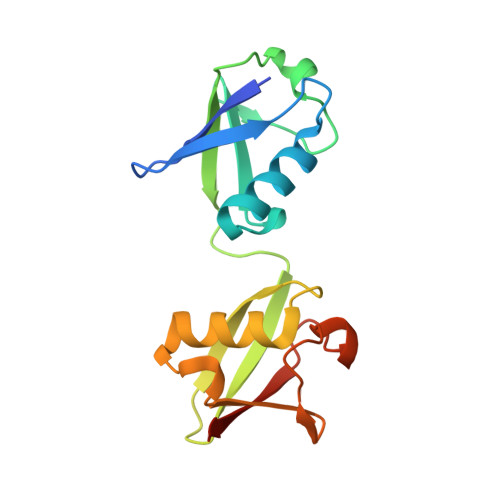
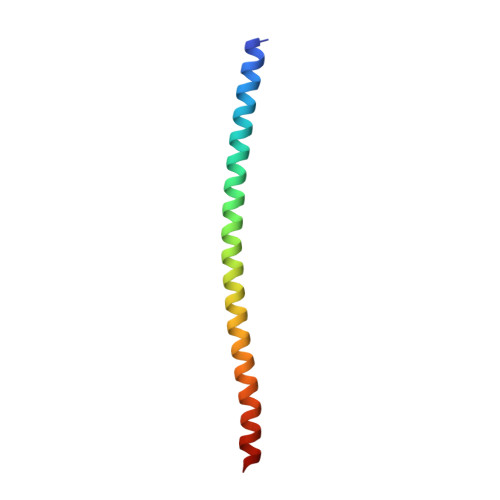Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
Herhaus, L., van den Bedem, H., Tang, S., Maslennikov, I., Wakatsuki, S., Dikic, I., Rahighi, S.(2019) J Mol Biology 431: 3146-3156
- PubMed: 31247202
- DOI: https://doi.org/10.1016/j.jmb.2019.06.012
- Primary Citation of Related Structures:
6N5M, 6N6R, 6N6S - PubMed Abstract:
Although the Ub-binding domain in ABIN proteins and NEMO (UBAN) is highly conserved, UBAN-containing proteins exhibit different Ub-binding properties, resulting in their diverse biological roles. Post-translational modifications further control UBAN domain specificity for poly-Ub chains. However, precisely, how the UBAN domain structurally confers such functional diversity remains poorly understood. Here we report crystal structures of ABIN-1 alone and in complex with one or two M1-linked di-Ub chains. ABIN-1 UBAN forms a homo-dimer that provides two symmetrical Ub-binding sites on either side of the coiled-coil structure. Moreover, crystal structures of ABIN1 UBAN in complex with di-Ub chains reveal a concentration-dependency of UBAN/di-Ub binding stoichiometry. Analysis of UBAN/M1-linked di-Ub binding characteristics indicates that phosphorylated S473 in OPTN and its corresponding phospho-mimetic residue in ABIN-1 (E484) are essential for high affinity interactions with M1-linked Ub chains. Also, a phospho-mimetic mutation of A303 in NEMO, corresponding to S473 of OPTN, increases binding affinity for M1-linked Ub chains. These findings are in line with the diverse physiological roles of UBAN domains, as phosphorylation of OPTN UBAN is required to enhance its binding to Ub during mitophagy.
- Institute of Biochemistry II, Goethe University School of Medicine, Theodor-Stern-Kai 7, 60590 Frankfurt am Main, Germany.
Organizational Affiliation: